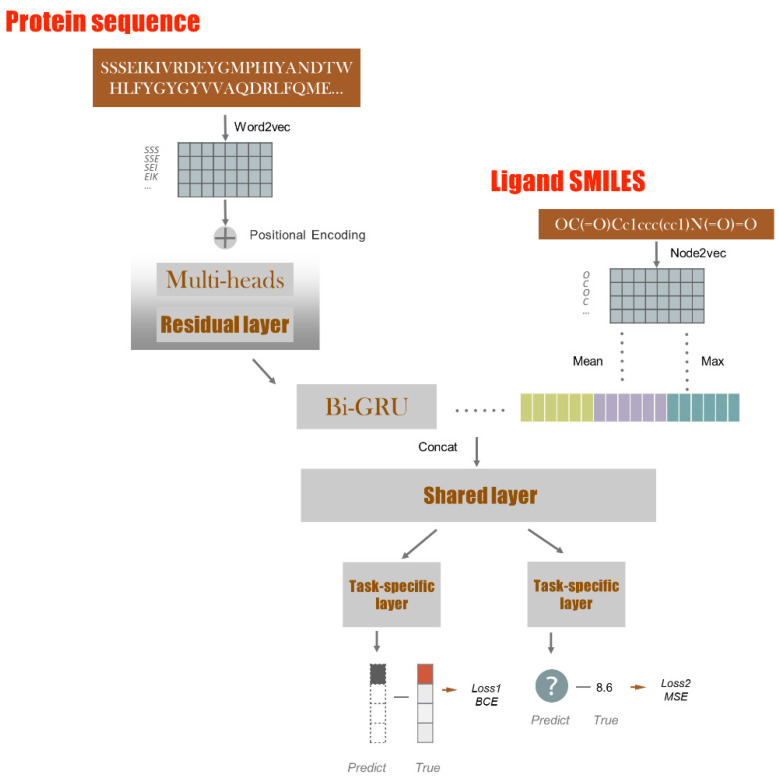
Figure 1.
Schematic of the proposed model. The model involves two parts: protein/ligand feature extraction and their interaction prediction. First, the protein sequence is processed in turn by word2vec [13], multi-heads residual layer and Bi-GRU (bidirectional gated recurrent unit) modules. Ligand smiles is processed by node2vec [14]. Then, their representations are fed into a shared layer and task-specific layer and connected to the labels.