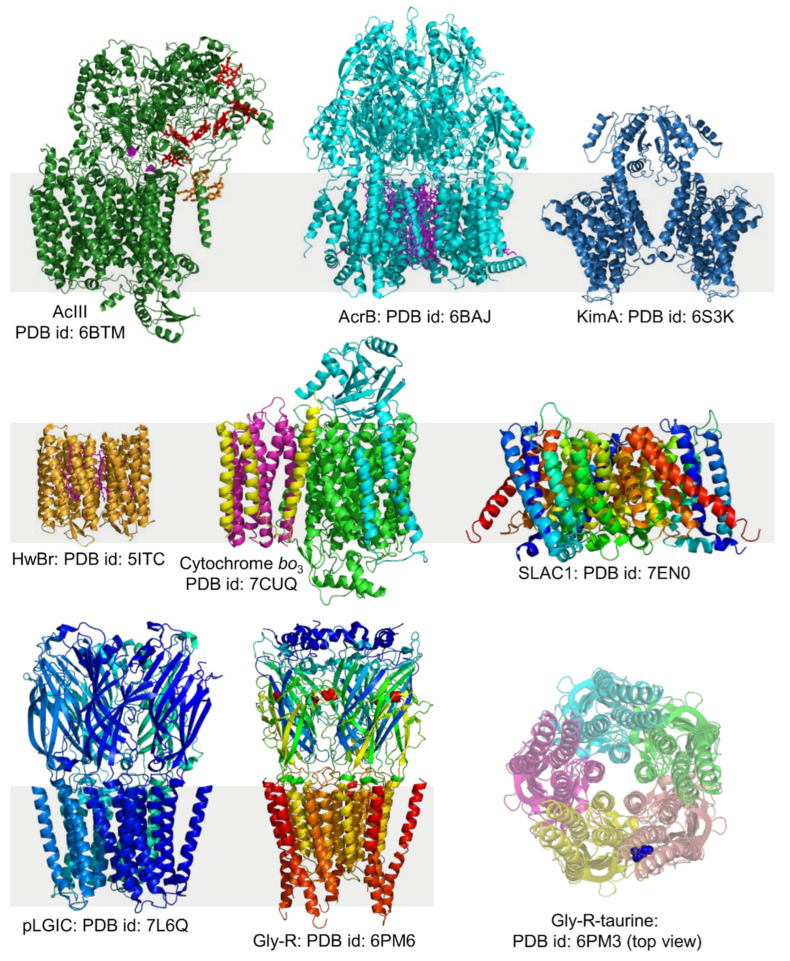
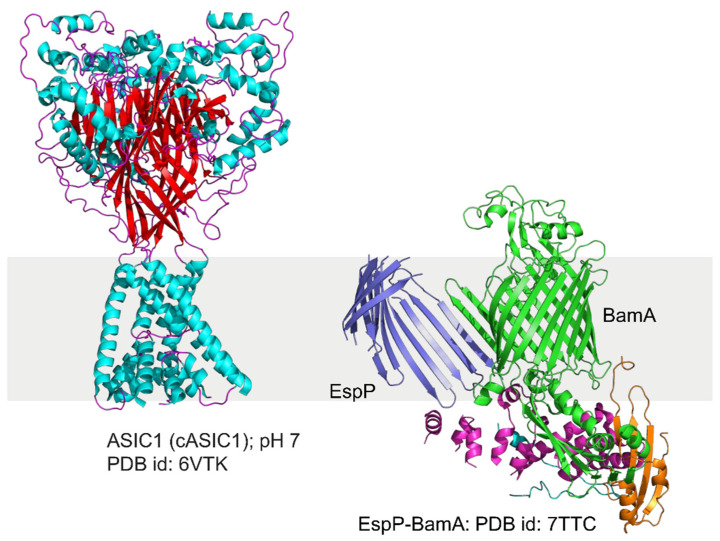
Figure 8.
The structure of alternative Complex III (PDB id: 6BTM) from Flavobacterium johnsoniae [184], AcrB (PDB id: 6BAJ) from E. coli (K-12) [182], KimA (PDB id: 6S3K) from Bacillus subtilis [157], HwBr (PDB id: 5ITC) from Haloquadratum walsbyi [119], cytochrome bo3 (PDB id: 7CUQ) from E. coli [171], SLAC1 (PDB id: 7EN0) from Brachypodium distachyon SLAC1 [173], bacterial pLGIC (PDB id: 7L6Q) [172], the glycine receptor open conformation (PDB id: 6PM6; the Gly residues are shown in red spheres) and taurine-bound closed conformation (top view) (PDB id: 6PM3; taurine is shown in blue spheres) from zebrafish [185]. The 6PM3 structure is shown with 50% transparency to highlight taurine. ASIC1 (PDB id: 6VTK) from chicken [186], and EspP-BamA complex structure (PDB id: 7TTC) from E. coli [136]. The structures were generated using PyMOL.