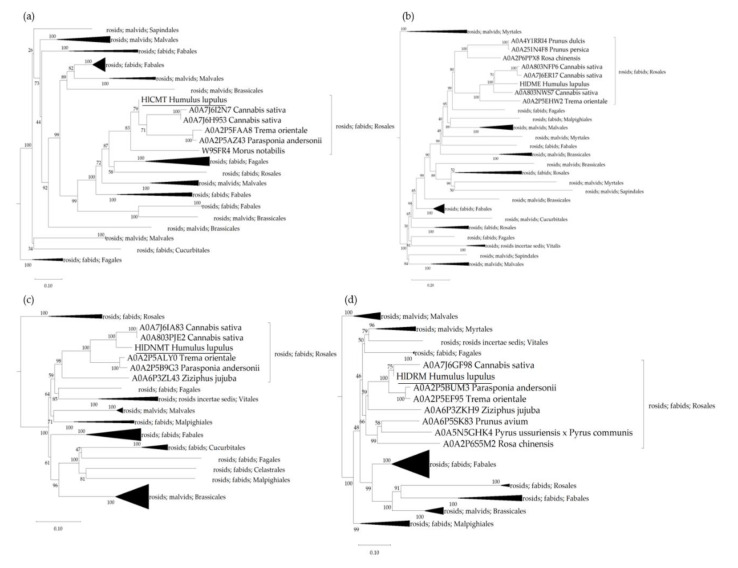
Figure 3.
Phylogenetic trees of the Chromomethylase (CMT), Demethylase (DME), DNA methyltransferase (DNMT), and Domains Rearranged Methyltransferase (DRM) proteins for the selected plant species (Supplementary Table S2), containing the newly identified DNA methylases and demethylases in hop. The polypeptide sequences were aligned using the MUSCLE algorithm, and the maximum likelihood phylogeny based on the WAG protein substitution model was used to construct the neighbour-joining trees for the (a) CMT, (b) DME, (c) DNMT, and (d) DRM protein groups. The numbers above the nodes indicate the reliability of 1000 bootstrap replicates. Scale bars represent amino acid substitutions per site. The software MEGA11 was used for the visualisation of the phylogenetic trees [54].