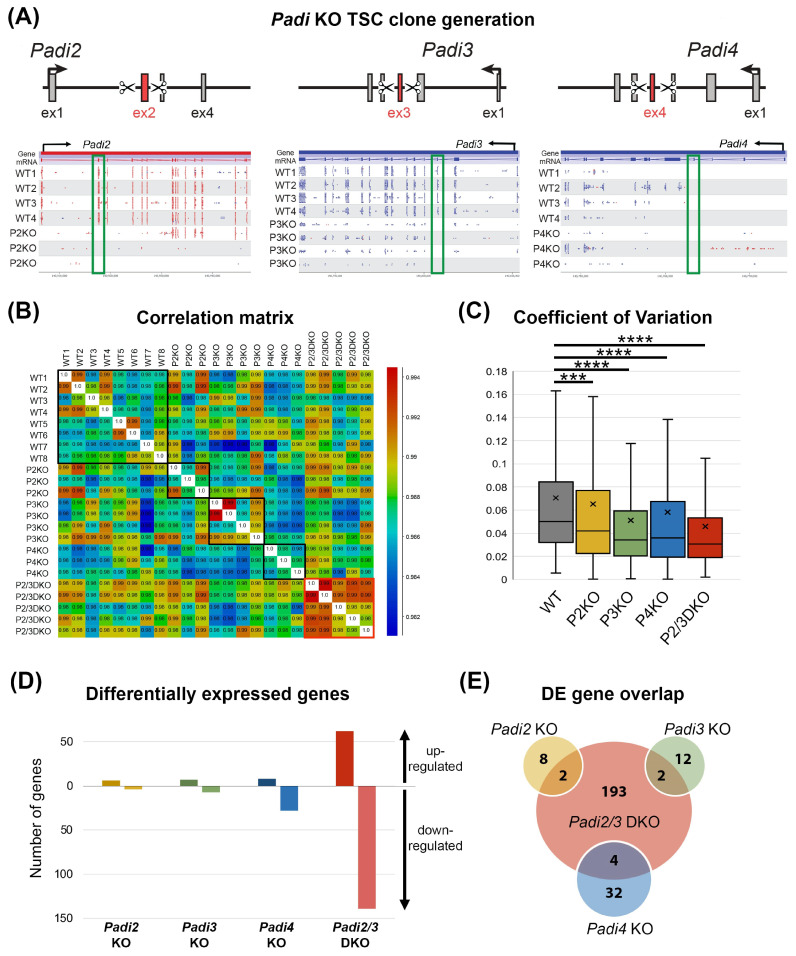
Figure 2.
Padi knockout (KO) TSC generation and transcriptomic analysis. (A) Depiction of the Padi genes with the exons targeted for deletion by CRISPR/Cas9-mediated knockout highlighted in red. Deletion of the targeted exons results in frameshift mutations and premature translational termination. SeqMonk screenshots of RNA-seq data of wild-type (WT) and independently derived Padi2 (P2)-, Padi3 (P3) and Padi4 (P4)-KO clones across the relevant genomic regions are shown underneath, with the targeted exons highlighted by the green rectangle. A complete absence of reads across the deleted exons provides an independent confirmation of correct gene ablation. Of note, Padi4 exon 4 expression was detectable at low levels in some WT TSCs. (B) Correlation matrix of global gene expression profiles across all TSC clones of the indicated genotypes. Rectangles highlight the relevant samples comparing inter-clone variability in each genotype. Padi2/3 double KO (DKO) clones are the most similar to each other, whereas WT clones display a higher extent of clonal variability. (C) The coefficient of variation of the top 10,000 expressed genes in WT and Padi KO/DKO clones. All Padi KOs exhibit reduced transcriptional heterogeneity. This effect is most pronounced in the Padi2/3 DKO TSCs. Statistical analysis was conducted by one-way ANOVA with Dunnett’s multiple comparisons test. *** p < 0.001, **** p < 0.0001. (D) Diagram of up- and down-regulated genes in the various KO/DKO TSC clones as determined by DESeq2 analysis (p < 0.05). (E) Overlap of differentially expressed (DE) genes in the various Padi KO/DKO TSCs.