Abstract
Acute myeloid leukemia (AML) is an aggressive malignancy of the bone marrow with 5-year overall survival of less than 10% in patients over the age of 65. Limited progress has been made in the patient outcome because of the inability to selectively eradicate the leukemic stem cells (LSC) driving the refractory and relapsed disease. Herein, we investigated the role of the reprogramming factor KLF4 in AML because of its critical role in the self-renewal and stemness of embryonic and cancer stem cells. Using a conditional Cre-lox Klf4 deletion system and the MLL-AF9 retroviral mouse model, we demonstrated that loss-of-KLF4 does not significantly affect the induction of leukemia but markedly decreased the frequency of LSCs evaluated in limiting-dose transplantation studies. Loss of KLF4 in leukemic granulocyte-macrophage progenitors (L-GMP), a population enriched for AML LSCs, showed lessened clonogenicity and percentage in the G2/M phase of the cell cycle. RNAseq analysis of purified L-GMPs revealed decreased expression of stemness genes and MLL-target genes and upregulation of the RNA sensing helicase DDX58. However, silencing of DDX58 in KLF4 knockout leukemia indicated that DDX58 is not mediating this phenotype. CRISPR/Cas9 deletion of KLF4 in MOLM13 cell line and AML patient-derived xenograft cells showed impaired expansion in vitro and in vivo associated with a defective G2/M checkpoint. Collectively, our data suggest a mechanism in which KLF4 promotes leukemia progression by establishing a gene expression profile in AML LSCs supporting cell division and stemness.
Keywords: leukemic stem cells, KLF4, MLL-AF9, transcription factor
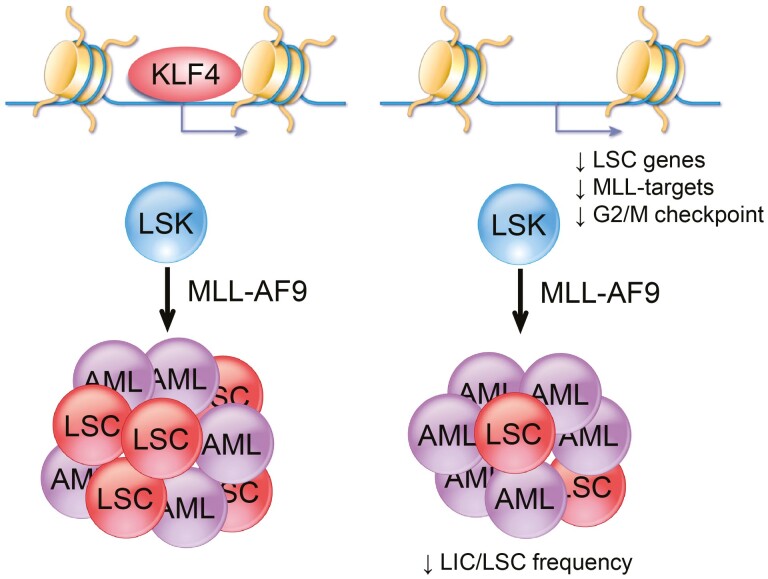
Graphical Abstract
KLF4 promotes leukemic stem cell survival in MLL-AF9 acute myeloid leukemia by regulating genes in the leukemic stem cell signature, MLL targets, and cell cycle.
Significance Statement.
Many elderly patients who have acute myeloid leukemia (AML) fail to respond to treatment and develop the refractory and relapsed disease, and thus alternative therapies are needed to eradicate chemoresistant cells. This study identified a new function for the reprogramming factor KLF4 in leukemic stem cell (LSC) maintenance, a rare leukemic population responsible for treatment failure in AML. KLF4 regulates the expression of genes involved in leukemia stem cells, MLL-AF9 targets, and the cell cycle. Identifying molecular mechanisms supporting LSC will pave the way for the future development of alternative therapies.
Introduction
Acute myeloid leukemia (AML) is a rapidly progressing hematological malignancy featuring clonal expansion and accumulation of immature myeloid progenitor cells with a differentiation block.1 While other leukemias have seen significant improvements in clinical outcomes through the past 3 decades, AML has lagged with 25% of patients responding to the standard of care mainly due to the disease’s genetic heterogeneity, the frailty of the patient population (commonly over 65 years of age), and high frequency of refractory and relapsed disease.2 Leukemic stem cells (LSCs) are rare leukemic populations that maintain the disease and are responsible for chemoresistance and disease relapse.3-5 Thus, a better understanding of how this population is maintained will provide a molecular framework to develop alternative therapies.
The immunophenotypic identification of AML LSCs has allowed research studies to attempt the development of targeted therapy and revealed a complex picture in which leukemic cells with stem-like properties share normal stem and myeloid progenitor cell surface markers.6 The progressive accumulation of genetic mutations in LSCs and clonal evolution create LSCs unique to each patient, making it challenging to identify molecular targets for developing LSC-directed therapy. As the immunophenotypic classification of AML LSCs in patients has been limited by the reality that LSC properties can be found in cells with diverse surface immunophenotypes,6 identifying critical molecular signaling pathways required for leukemia maintenance is an alternative approach applicable to LSCs independently of the immunophenotype and genetic background of AML. This approach has led to the identification of non-genetic prognostic indicators, such as the 17-gene stemness score capable of accurately stratifying patient outcomes7 and the study of individual transcriptional signaling mechanisms within the LSC, including WNT/β-Catenin,8-10 BMI1,11,12 and C/EBPα.13,14MLL-rearranged AMLs represent a poor prognosis subset usually presenting as FAB-M4/M5.15,16 This AML subtype relies on aberrantly expressed embryonic stemness gene expression programs—notably HOX family genes activated directly by MLL-fusion proteins17 and MYC, which is frequently upregulated through super-enhancers.18,19 Less is known about the importance of other critical transcriptional regulators in supporting the MLL-rearranged LSC.
The Krüppel-like factor 4 (KLF4) is a zinc-finger transcription factor that regulates self-renewal in embryonic stem cells and contributes to the reprogramming of somatic cells into induced pluripotent stem cells (iPSC).20 KLF4 controls gene regulation through various mechanisms, including DNA binding and recruitment of co-activators and co-repressors such as p300 and HDACs,21-23 organizing higher chromatin structures,24,25 and protein-protein interactions. These regulatory mechanisms have been associated with the control of cell cycle progression, apoptosis, differentiation, and self-renewal in developmental, immunological, and cancerous contexts. In normal hematopoiesis, our group and others reported that KLF4 promotes monocyte development in murine in vivo models lacking KLF4,26,27 controls proliferation in naïve CD8+ T-cells,28 and supports the survival of NK cells.27 In examining hematologic malignancies, there are cases in which KLF4 expression is reported to contribute to apoptosis and overall tumor-suppressive function, as in Hodgkin lymphoma,29 chronic lymphocytic leukemia,30 and T-cell acute lymphoblastic leukemia.31,32 However, KLF4 can also have a pro-oncogenic function. For example, we reported that KLF4 supports LSC self-renewal in BCR-ABL1 induced chronic myeloid leukemia (CML) through repression of the kinase DYRK2.33 These findings revealed that KLF4 function in different leukemias depends on oncogenic drivers and activated cellular pathways. Studies in bulk AML leukemic cells have reported a correlation of KLF4 expression levels with morphologic differentiation,34 and induction of monocytic differentiation and apoptosis upon overexpression of KLF4 in AML cell lines.22,35-38 These findings have led to an assertion that KLF4 exerts a tumor-suppressive function in AML. However, the induction of KLF4 has been reported in the ZMYM2-FGFR1-driven leukemogenesis.39 In addition, using CRISPR/Cas9-mediated deletion, we have reported that KLF4 expression supports cell growth and survival in the AML cell lines.40MLL-rearranged AMLs, a particularly abysmal prognosis cytogenetic category, express elevated KLF4 levels.37,41 However, whether this expression plays a role in supporting MLL-rearranged AML LSCs has not been previously described.
Here, we demonstrate that KLF4 supports MLL-AF9-driven AML while maintaining the expression of genes involved in the stemness of LSCs and MLL target genes. Loss of KLF4 reduces LSC frequency and disease progression in a murine AML model and CRISPR-Cas9-edited patient-derived AML xenografts. L-GMPs from KLF4 knockout leukemias demonstrated defects in clonogenicity and cell cycle progression. At the molecular level, the loss of KLF4 in LSCs results in the disruption of gene sets associated with stem cells, MLL-AF9 targets, and upregulation of the RNA sensing helicase DDX58. In the human MA9 positive cell line MOLM13, deletion of KLF4 reduced expansion both in vitro and in vivo associated with a defective G2/M checkpoint. Our data reveal a pro-leukemic function of KLF4 in AML by maintaining the pool of LSCs, providing potential molecular targets for developing targeted therapy.
Materials and Methods
MLL-AF9 Mouse Model
For primary leukemias, lineage− Sca-1+ c-Kit+ (LSK) cells were purified from the bone marrow of 8–12-week-old Klf4fl/fl, Klf4fl/fl Vav-Cre, or Klf4fl/fl ROSA-ERCre mice (Fig. 2 only) using hematopoietic stem and progenitor cell enrichment kit (BD BiosciencesTM) and cell sorting. LSK cells were plated in X-Vivo (Lonza) supplemented with murine IL-3 (10 ng/mL), SCF (100 ng/mL), and human IL-6 (6 ng/mL) (PeproTech) overnight to stimulate proliferation. LSK cells were transduced with pMIG-MLL-AF9-3xty1 (provided by Dr Daisuke Nakada, Baylor College of Medicine) in 2 spinoculations using retronectin (Takara)-coated plates. Twenty-four hours after the second spinoculation, 10 000 cells containing ~10% transduced cells were mixed with 200 000 radioprotective bone marrow cells and transplanted to lethally irradiated (950 rads) C57BL/6 recipient mice. Engraftment was confirmed by the detection of GFP+ CD11b+ cells in peripheral blood. For survival experiments, mice were deemed moribund and euthanized when they lost more than 20% of body weight or showed signs of disease such as hunched posture, reduced motility, and pale skin.
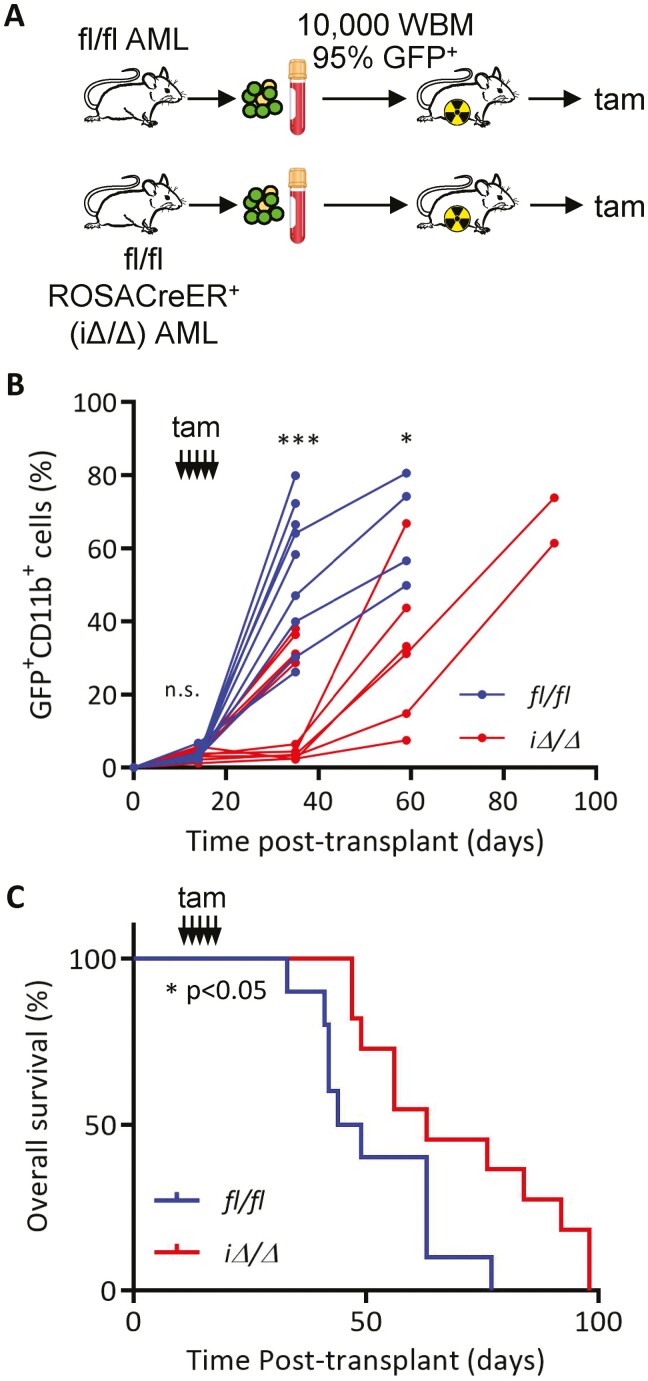
Figure 2.
Post-transplant deletion of KLF4 delays leukemia progression. (A) Schematic diagram of induction of Klf4 deletion using the ROSA-CreER system. Cryopreserved leukemic cells were co-injected with whole bone marrow (WBM) in ablated mice. (B) Flow cytometric analysis of GFP+ CD11b+ cells in peripheral blood of fl/fl (n = 10) and iΔ/Δ (n = 11) MA9 mice after induction of gene deletion via daily administration of tamoxifen (Tam) beginning at day 14 post-transplantation and continuing for 5 days. (C) Kaplan–Meier analysis of survival of fl/fl (n = 10) and iΔ/Δ (n = 11) MA9 mice. *P < .05, ***P < .001. Two-tailed Student’s t test was used in B. Log-rank test was used in C.
Monitoring of Peripheral Blood Cells
Peripheral blood was collected by tail vein prick. Red blood cells (RBCs) were lysed by hypotonic incubation with water. The remaining white blood cells were treated with anti-mouse CD16/32 antibody (Fc Block) (Biolegend) for 5 min, followed by staining with anti-mouse CD11b—PE-Cy7 antibody (eBioScience), Gr-1—APC antibody (Biolegend), TCRβ—PE antibody (Biolegend), and CD19—PerCP-Cy5.5 antibody (Biolegend) at 4 °C for 30 min. Blood cells were washed with PBS and analyzed using the FACSCanto instrument (BD) and FlowJo software. For complete blood count (CBC) analysis, 15 μL of blood was analyzed using the Heska HT5 Hemocounter. For blood smears, 2 μL of whole blood was added to glass slides and streaked using Hemaprep (J.P. Gilbert) and stained with Hema 3 Stat Pak (Fisherbrand).
Cryopreservation of MA9-Induced Leukemic Bone Marrow
Mice were euthanized by CO2 inhalation and secondary bilateral opening of the thorax. Bone marrow cells were flushed from the femur and tibia using PBS with 3% FBS and counted using Cellometer Auto 2000 (Nexcelom). The spleen was weighed, and the cell suspension was prepared using a plunger and 40 μm cell strainer, flushing with PBS with 3% FBS, and RBCs were lysed. Samples were analyzed by flow cytometry to confirm myeloid leukemia burden and frozen in X-vivo with 10% DMSO. Diseased mice showed ~95% GFP-positive cells in the bone marrow.
Murine Colony-Forming Cell Assay
Leukemic MA9 cells were counted and plated at 5000 cells/well in 1 mL MethoCult GF M3434 (STEMCELL Technologies) in a 12-well plate. For replating of L-GMP, 1000 cells were plated in the first plating and 250 in the second plating. Colonies were counted after each week.
In Vivo Tamoxifen-Induced Deletion of Klf4
A total of 10 000 cells from primary Klf4fl/fl MA9 AMLs or Klf4fl/fl ROSA-ERCre MA9 AMLs (before any tamoxifen exposure) were transplanted (tail vein injection) into sublethally irradiated (600 rad) recipients. Detection of GFP+ cells in peripheral blood was used to confirm engraftment on day 12. Tamoxifen (0.125 mg intraperitoneally) was administered daily for 5 consecutive days starting on day 14 to induce Klf4 deletion in ROSA-ERCre recipients (iΔ/Δ). Klf4fl/fl served as tamoxifen-receiving control. Monitoring was conducted as stated above.
Leukemia Initiating Cell Limiting Dose Cell Transplantation
MA9 primary AML cryovials were thawed and live cells were counted. From each vial, cells were diluted to 50 000, 25 000, 10 000, 5000, and 1000 cells per 200 µL in sterile saline solution and transplanted into sublethally irradiated (600 rad) recipients. The resulting disease penetrance was used to calculate leukemia-initiating cell frequency using the L-Calc software tool available from STEMCELL Technologies.
Isolation of L-GMP
A total of 50 000 viable cells were transplanted to sublethally irradiated (600 rad) recipients, and bone marrow cells were collected by flushing after 3–4 weeks post-transplant. Cells were enriched using hematopoietic stem and progenitor cell enrichment kit (BD BiosciencesTM) and stained for L-GMP surface markers (c-Kit PE eBioScience, Sca-1 PE-Cy7 eBioScience, CD16/32 FcR APC-Cy7 BD Pharmingen, CD34 APC BioLegend) with L-GMP considered GFP+lin−Sca-1−c-Kit+CD34+CD16/32FcR+. >95% of GFP+lin−Sca-1−c-Kit+ cells were CD34+CD16/32FcR+.
Cell Cycle Analysis of L-GMPs
For cell cycle analysis, freshly sorted L-GMPs and L-GMPs cultured in X-Vivo with murine IL-3 (10 ng/mL), SCF (100 ng/mL), and human IL-6 (6 ng/mL) (PeproTech) were lysed in hypotonic DNA-staining solution containing 0.1% sodium citrate, 0.1% Triton-X, 100 μg/mL RNAse A, 50 μg/mL propidium iodide in de-ionized water. All samples were plated in triplicate. Cells were analyzed in a BD FACSCanto instrument and Watson (Pragmatic) model in the FlowJo software to calculate cell cycle phases.
Cell Lines and Cell Culture
MOLM13 cells were obtained from Dr Rachel Rau (Baylor College of Medicine). MOLM13 cells were maintained in RPMI 1640 medium (Lonza) containing 10% fetal bovine serum (FBS), 2 mM L-glutamine. The cell line was authenticated every 6 months (Cell Line Characterization, MD. Anderson) and periodically tested for mycoplasma.
Generation of MA9 PDX Line and CRISPR/Cas9 Editing of KLF4
AML cells (peripheral blood) harboring the MLL-AF9 translocation from a 64-year-old female patient were purchased from the Fred Hutchinson repository. PDX line was generated according to the ProXe protocol42 and characterized for myeloid surface marker expression by flow cytometry. To delete KLF4 in AML PDX cells, human CD45+ cells were purified from P1 NSG-SGM3 mouse using human-mouse chimera isolation kit (STEMCELL Technologies), and 5M cells were nucleofected with multiplex sgRNAs (Synthego) targeting human KLF4 (GCCATGTCAGACTCGCCAGG, CGCCGGGCCAGACGCGAACG, and GAGCGATACTCACGTTATTC) and Cas9 nuclease using the Neon Transfection System (ThermoFisher) with 1400 V, 3 pulses, 10 ms in 100 µL tip. A total of 500 000 cells were transplanted per mouse into NSG-SGM3 mice, which had received 250 rads irradiation. KLF4 deletion was assessed by sequencing genomic DNA from peripheral blood (isolated by QIAGEN DNeasy Blood and Tissue Kit) after PCR amplification using forward primer GTGTTATGTCCTGTCTGCCCAATT and reverse primer GTTTTGGCTTCGTTTCTTCTCTTC, spanning the Cas9-sgRNA cleavage site. PCR amplicons were then used for sequencing analysis using sequencing primer CTTACCCTCGTTCAGTGGCTCTT to identify the knockout efficiency using Inference of CRISPR Edits (ICE) from Synthego. Expansion of AML PDX cells was monitored by flow cytometric detection of human CD45-expressing cells in peripheral blood.
RNASeq Analysis of L-GMPs
A total of 50 000–300 000 L-GMPs were submitted in RNALater solution to UCLA Technology Center for Genomics & Bioinformatics (TCGB) Laboratory for RNASeq. Fastq files were processed using Partek Flow software to trim bases, align reads (STAR), quantify, normalize, and account for sample batch effect. Genes were filtered for low expression with a 10 normalized counts cut-off. The 3D PCA was produced using the pca3d function and prcomp functions in R.43,44 For volcano plot and significantly altered genes, P-values were evaluated with 2-tail heteroscedastic t-tests in Microsoft Excel 2016. FDR was adjusted via a Benjamini–Hochberg correction and genes with passing a threshold of FDR lower than 0.1 were considered. For GSEA, Gene set enrichment calculations were completed using GSEA (v. 4.0.3) (PMID: 16199517) with statistics estimated by 10K gene set permutations. The Broad-UC San Diego Molecular Signatures Database (MSigDB v7.0) curated and Hallmark (PMID: 26771021) pathway sets were used, along with the v7.0 mapping to human orthologs. Venn diagram comparing murine fl/fl and Δ/Δ HSC & L-GMP, combined with human L-GMP overlap, was prepared by processing L-GMP and HSC FASTQ files through a pipeline of Trimmomatic, STAR, RSubreadfeatureCounts, RawCounts, and DEseq2 with log2fold≥1, pAdjust<0.05 and plotted in R. Human GMP and L-GMP comparison was performed from data available in GSE35008.
Quantitative Real-time PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen) from murine AML bone marrow. cDNA was synthesized from 100 to 500 ng RNA using oligoDT primers and a SuperScript III kit (Invitrogen). PowerUp SYBR Green Master Mix (Applied Biosystems) was used for quantitative real-time PCR as specified by the manufacturer. Reactions were performed using the StepOnePlus Real-Time PCR System (Applied Biosystems). Relative expression was normalized to β-actin expression (ΔΔCT). The following primers were used:
| mouse KLF4 | Forward | TTTCCAACTCGCTAACCCACCA |
| Reverse | TCATTGATGTCCGCCAGGTTGA | |
| mouse β-actin | Forward | GTGGGCCGCTCTAGGCACCA |
| Reverse | CGGTTGGCCTTAGGGTTCAGGGGGG | |
| mouse DDX58 | Forward | AGCCAAGGATGTCTCCGAGGAA |
| Reverse | ACACTGAGCACGCTTTGTGGAC | |
| mouse PBX3 | Forward | ACCCTCCGTCATGTTATCAATC |
| Reverse | CCAGCCTCCATTAGCGTTTA | |
| mouse DACH1 | Forward | TCTAACTGGGCATGGACAAC |
| Reverse | GGCCCTGTATGTTAGTGAGAAG | |
| mouse GAS7 | Forward | GATGACACTTCCTCCAGCATAA |
| Reverse | GGAAAGGCATGTGTGGATAGA | |
| mouse ITGB3 | Forward | GGAATAGAACCCAGGACATCAC |
| Reverse | CCGTATTTACTCTCGGCATCTT | |
| mouse Meis1 | Forward | TCACACTGCTGGAGACGCAA |
| Reverse | ATCGTGGAGGGGATGCCTAC | |
| mouse Hoxa9 | Forward | CGTGACTGTCCCACGCTTGA |
| Reverse | TAGGGGCATCGCTTCTTCCG |
KLF4 ChIP PCR in DDX58 Gene
Three million fl/fl MA9 L-GMP cells were processed for cross-linking and immunoprecipitation according to ChIP-IT High Sensitivity Kit Protocol (Active Motif). KLF4 (Santa Cruz Biotechnology, sc-20691X) and IgG (Cell Signaling #2729S) were used for immunoprecipitation of chromatin. Primers targeting the DDX58 promoter from +365 to +464 (F-TGTGTAACCCGGTGTATGTG; R-CAGCTTGCAGGAATTTTGTT) were used to amplify immunoprecipitated chromatin and input (control).
Immunoblot Analysis
Cell pellets were lysed with SDS lysis buffer (1% SDS, 10 mM Tris pH 7.4, 1 mM phenylmethylsulfonyl fluoride) and supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher). Protein samples were resolved by SDS–PAGE (Novex NuPage Bis Tris Gel; Invitrogen MiniGel Tank) and then transferred onto PVDF membranes (iBlot 2 system, Invitrogen). The following antibodies were used: DDX58 (Rig-I (D14G6) Rabbit mAb #3743), Akt (Cell Signaling #9272), p-Akt T308 (Cell Signaling #13038P), S6K (Cell Signaling #2708P), p-S6K T389 (Cell Signaling #9205S), Direct-Blot HRP anti-β-actin antibody, PARP antibody from Cell Signaling. Primary antibodies were used at 1:1000 dilution and HRP anti-β-actin at 1:300 000. HRP cross-linked secondary antibodies (anti-rabbit IgG, HRP-linked antibody #7074 and anti-mouse IgG, HRP-linked antibody #7076 from Cell Signaling) were detected by West Femto Maximum Sensitivity Substrate (Thermo Fisher) and Amersham Hyperfilm ECL (GE). All secondary antibodies are used at 1:15 000–1:30 000 dilution. Protein quantity was normalized based on housekeeping control bands.
shRNA Knockdown of DDX58 in Δ/Δ MA9 AML
LSK cells from Δ/Δ mice were transduced with retrovirus MIG-MA93xty1-mNeptune and DDX58-targeted GFP lentivirus or scrambled sequence targeted control from Origene (TR30037 Ddx58 Mouse shRNA Plasmid-Locus ID 230073). Dual-transduced cells were transplanted to lethally irradiated (950 rad) recipient mice. After 5 weeks, mice were euthanized and GFP+mNeptune+ cells were sorted using FACSAria Instrument (BD Biosciences). A total of 12 500 cells were transplanted per recipient to sub-lethally irradiated recipients (600 rad). The expansion of DDX58 shRNA and scr shRNA-expressing AML cells was monitored by detecting GFP+mNeptune+ cells in peripheral blood. Cells were also plated in methylcellulose for quantitation of colony-forming units. Cells from the CFU assay were used to assess the efficiency of DDX58 knockdown by immunoblot.
Generation of MOLM13 KLF4 KO Clonal Lines
To knockout the KLF4 gene in MOLM13 cells, we used 3 chemically modified synthetic sgRNAs from Synthego with sequences GCCATGTCAGACTCGCCAGG, CGCCGGGCCAGACGCGAACG, and GAGCGATACTCACGTTATTC. We followed the instructions obtained from the manufacturer to form the RNP complex with Cas9 (Cas9 plus sgRNAs). Briefly, the Cas9-sgRNA RNP in a total volume of 12 μL were electroporated using the Neon transfection system (Thermo Fisher Scientific) under the following conditions: 1400 V, 10 ms, and 3 pulses. Electroporated cells were cultured in a growth medium for 3 days, and then single-cell clones were obtained by diluting cells to 0.5 cells/100 μL and plating in a 96-well U-bottom plate for 2 weeks. Genomic DNAs were isolated and then used for PCR amplification using forward primer GTGTTATGTCCTGTCTGCCCAATT and reverse primer GTTTTGGCTTCGTTTCTTCTCTTC, spanning the Cas9-sgRNA cleavage site. PCR amplicons were then used for sequencing analysis using sequencing primer (CTTACCCTCGTTCAGTGGCTCTT) to identify the knockout efficiency using Inference of CRISPR Edits (ICE), which is a free and open-source software tool that offers a fast and reliable analysis of CRISPR editing data from Synthego. Knockout was additionally verified by immunoblotting and PCR detection of cleaved genomic DNA on 2.0% agarose gel. Two knockout clones were selected for further study.
Cell Growth and Cell Cycle Assays
For analysis of cell growth, MOLM13, KLF4ko1, KLF4ko2 cells were plated at 100 000 cells/ml and counted in 24-h intervals. For cell cycle analysis, MOLM13, KLF4ko1, KLF4ko2 cells were plated at 100 000 cells/mL, collected after 24 h, and lysed in hypotonic DNA-staining solution containing 0.1% sodium citrate, 0.1% Triton-X, 100 μg/mL RNAse A, 50 μg/mL propidium iodide in de-ionized water. All samples were plated in triplicate. Cells were analyzed in a BD FACSCanto instrument and Watson (Pragmatic) model in FlowJo software.
AML Cell Line Xenografts
MOLM13 cells were washed with PBS and resuspended in medical-grade physiologic saline solution. Cells (10 000) were injected into 10–20-week-old NOD SCID gamma (NSG) mice via tail vein. Mice were monitored in peripheral blood by cytometric detection of human CD45 to determine leukemic cell engraftment and disease progression. Mice were euthanized upon signs of moribundity that presented as hind-limb paralysis.
Reverse Phase Protein Array
Cell lysates, serial dilution of standards, and positive and negative controls were arrayed on nitrocellulose-coated slides (Grace Bio-Labs) by the Quanteriz 2470 Arrayer. Each slide was probed with a validated primary antibody plus a biotin-conjugated secondary antibody. Signal detection was amplified using an Agilent GenPoint staining platform and visualized by DAP colorimetric reaction. The slides were scanned, analyzed, and quantified using customized software (Array-Pro Analyzer, Media Cybernetics) to generate spot intensity. Each dilution curve was fitted with a logistic model (RPPA SPACE developed at MD Anderson). The protein concentrations were then normalized for protein loading. The corrections factor was calculated and normalized across sets by replicates-based normalization using an invariant set of control samples to adjust for batch differences between identical controls.45
Homing Assay
MLL-AF9 AML cells isolated from diseased mice were transplanted into ablated mice (800 rads) and bones were collected 18 h later for analysis. Bone marrow cells were analyzed for the expression of GFP to identify donor leukemic cells.
Statistical Analysis
Experiments were performed without blinding and with no exclusion of samples. Linear regression was performed with GraphPad Prism software. An unpaired 2-tailed Student’s t test was used for statistical analysis. The survival of leukemic mice was visualized using Kaplan–Meier curves, and statistical significance was calculated using the log-rank test (GraphPad Prism). P values were determined using GraphPad software. Results with a P-value < .05 were considered statistically significant.
Results
Loss-of-KLF4 Impairs Maintenance of MLL-AF9-Induced Leukemia
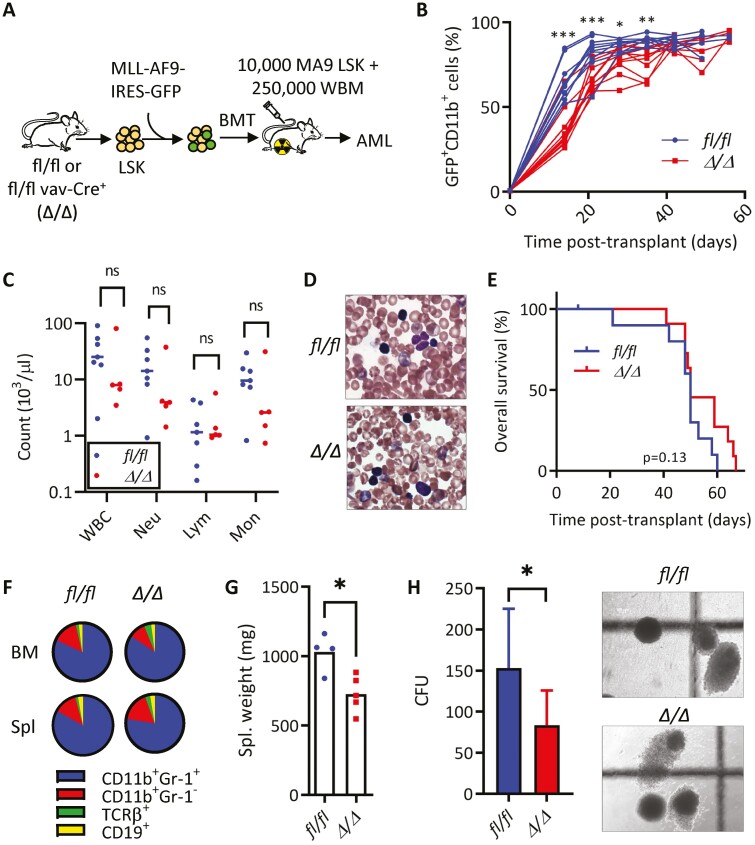
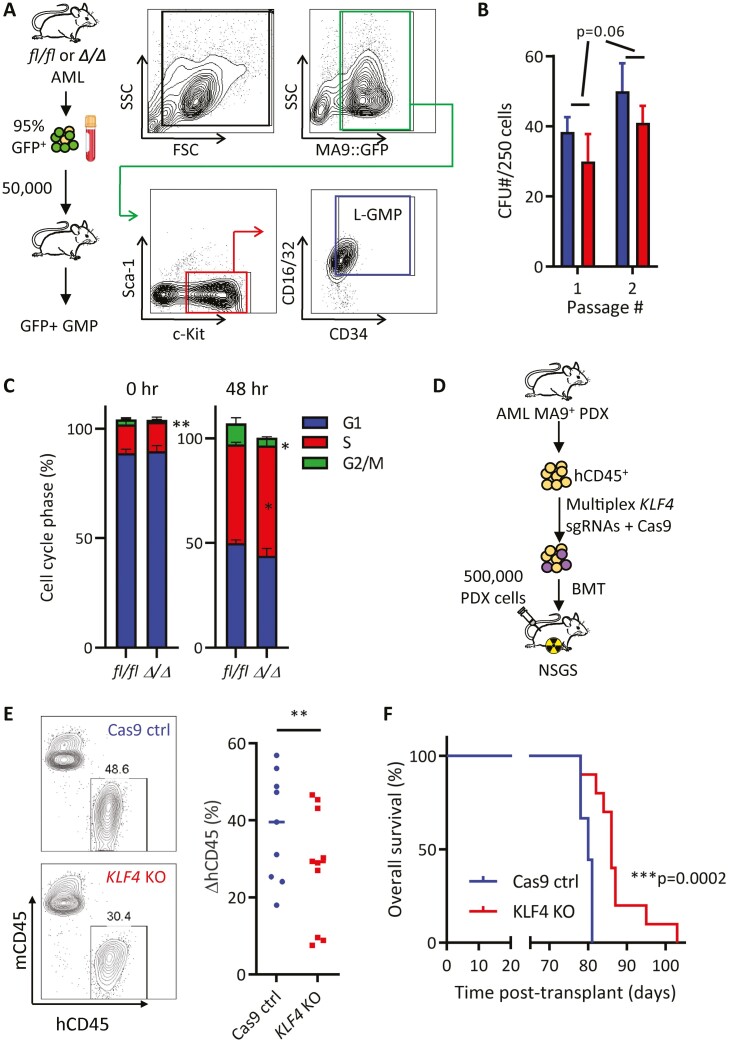
MLL-rearranged AMLs typically present as M4/M5 subtype disease as classified by the French-American British (FAB) classification system.16,41 Examining KLF4 in human disease, we observed elevated transcript levels in M4/M5 disease compared to other subtypes (Supplementary Fig. S1A), consistent with other analyses detecting high KLF4 levels in MLL- (11q23) rearranged AMLs.37 Further analysis of publicly available clinical data using the PrognoScan resource revealed AMLs with higher levels of KLF4 result in significantly worsened prognoses in multiple study cohorts (Supplementary Fig. S1B).46 Given these observations, we reasoned that deletion of the KLF4 gene in the context of the MLL-AF9 (MA9) oncogenic driver would provide a suitable model for studying the KLF4 function in AML. To evaluate the importance of KLF4 in a leukemic model which recapitulates the stem-cell and blast hierarchy similarly to human AML, we transformed Lin− Sca-1+ c-Kit+ (LSK) cells from Klf4fl/fl (fl/fl) and Klf4fl/flVav-Cre+(Δ/Δ) mice with the retrovirus pMIG-MLL-AF9-3xty1.47 A total of 10 000 cells were injected intravenously to lethally irradiated C57BL/6 mice and monitored for signs of leukemic development (Fig. 1A). A low GFP expression post-transduction prevented accurate transplantation based on the number of transduced cells. Myeloid leukemia cells (GFP+ CD11b+) expanded in the peripheral blood similarly in both groups, albeit less rapidly in Δ/Δ MA9 (leukemic) mice (Fig. 1B). Although the initial expansion of myeloid leukemia cells was reduced in the absence of KLF4, fl/fl and Δ/Δ mice showed similar disease burden as the disease progressed (Fig. 1B). We confirmed the successful deletion of Klf4 and induction of MA9 targets HOX and MEIS1 expression in AML cells by qPCR (Supplementary Fig. S2A-S2C). Post-mortem tissue analysis of moribund fl/fl and Δ/Δ MA9 (leukemic) mice showed similar morphological alterations in the spleen and infiltrations in the liver (Supplementary Fig. S2D). We did not detect significant differences in the number of white blood cells (WBC), neutrophils, and monocytes (Fig. 1C), as well as cells with blast-like morphology (Fig. 1D). This was reflected in a similar overall survival in MA9 Δ/Δ mice compared to fl/fl control in primary recipients (Fig. 1E). Analysis of moribund mice revealed ≥95% leukemic GFP+ myeloid cells (CD11b+ Gr-1+/CD11b+ Gr-1−) in the bone marrow of both groups and reduced spleen weight in MA9 Δ/Δ mice (Fig. 1F, 1G). Plating bone marrow cells from moribund mice in methylcellulose showed a reduction in colony formation in Δ/Δ leukemic cells (Fig. 1H).
Figure 1.
KLF4 is dispensable for the development of MLL-AF9-induced murine AML but supports disease progression. (A) Schematic diagram of MLL-AF9 (MA9)-induced leukemia using LSK cells from mice with conditional Klf4 deletion. (B) Flow cytometric detection of GFP+ CD11b+ leukemic cells in peripheral blood of fl/fl and Δ/Δ MA9 mice expressed as a percentage of total live cells (n = 10/group). (C) White blood cell counts at 5 weeks post-transplantation. (D) Representative blood smear of fl/fl and Δ/Δ MA9 mice at 7 weeks post-transplantation. (E) Kaplan–Meier analysis of survival of fl/fl and Δ/Δ MA9 mice (n = 10/group). (F) Frequency of myeloid (CD11b, Gr-1) and lymphoid (TCRβ, CD19) cells within GFP+ cells in the bone marrow and the spleen of moribund MA9 mice as determined by flow cytometry. A representative pie chart is shown. (G) Spleen weight of moribund fl/fl and Δ/Δ leukemic mice (n = 4/group). Mean and individual values are shown. (H) Colony-forming cell assay of fl/fl and Δ/Δ MA9 bone marrow cells from moribund mice (n = 7/group). The representative morphology of colonies is shown on the right. The data are represented as mean ± SD and are representative of 3 independent experiments. ns, not statistically significant. *P < .05, **P < .01, ***P < .001. Two-tailed Student’s t test was used in B, C, G, H. Log-rank test was used in E.
To evaluate whether loss of KLF4 post-engraftment altered MA9-induced leukemia, we transduced LSK cells from Klf4fl/fl and Klf4fl/flROSA-CreER (iΔ/Δ) mice and transplanted them into recipient mice. Leukemic cells were collected from diseased mice and cryopreserved for experiments. Primary leukemic cells (10 000) were transplanted into sub-lethally irradiated recipients, and Klf4 gene deletion was induced by tamoxifen administration on days 14–18 post-transplantation (Fig. 2A). MA9 iΔ/Δ mice showed a significant difference in the expansion of myeloid leukemia cells in the blood and improved overall survival (Fig. 2B, 2C). Altogether, these results indicate that loss of KLF4 impairs the expansion of leukemic cells and improves survival in MLL-AF9-induced AML.
KLF4 Regulates the Frequency of Leukemia-Initiating Cells in MLL-AF9-Driven AML
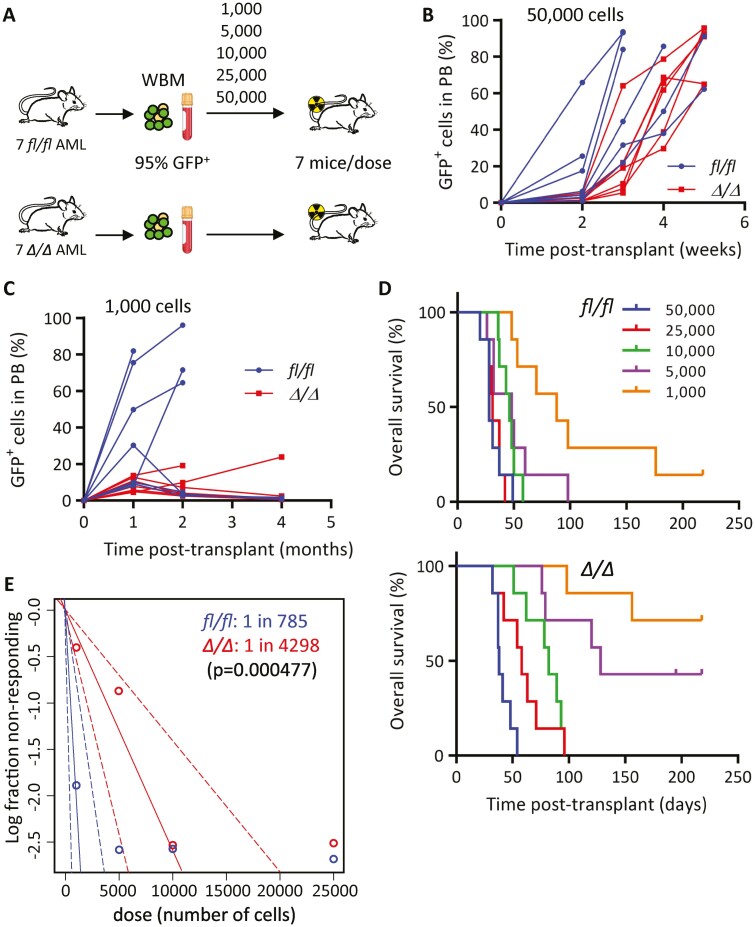
As KLF4 supports self-renewal and stemness in various cell subtypes, we next wanted to evaluate whether KLF4 plays a role in AML LSCs. The gold-standard assay for stem cell self-renewal is serial transplantation which assesses the capacity of stem cells to recapitulate leukemia. To determine the frequency of MA9 leukemia-initiating cells (LICs), we transplanted limiting doses of GFP+ bone marrow cells isolated from 7 independent leukemic mice for each group (fl/fl and Δ/Δ) into sub-lethally irradiated secondary recipients (Fig. 3A). In this case, sub-lethal irradiation was used instead of radioprotective bone marrow cells that could outcompete a small number of leukemic cells. Blood monitoring of mice transplanted with 50 000 and 1000 GFP+ bone marrow cells showed a significant delay in the expansion of Δ/Δ GFP+ (MA9) cells (Fig. 3B, 3C). Subsequently, the analysis of all transplanted doses showed prolonged latency, reduced disease penetrance, and significant improvement in survival in Δ/Δ MA9 AML mice (Fig. 3D). By evaluating the fraction of fully penetrant disease for each cell dose, we calculated a 5.5-fold lower LIC frequency present in Δ/Δ MA9 cells (Fig. 3E). We performed a homing/engraftment assay to rule out that a lower frequency was caused by impaired homing and found that Δ/Δ GFP+ (MA9) cells were able to home and engraft recipient bone marrow similarly to controls (Supplementary Fig. S3). We concluded that KLF4 contributes to sustaining the pool of LIC/LSCs in murine MA9-driven AML.
Figure 3.
KLF4 regulates the frequency of MLL-AF9 leukemia-initiating cells. (A) Schematic diagram of limiting-dose transplantation to enumerate leukemia-initiating cell frequency. Leukemic bone marrow cells from 7 independent leukemic MA9 mice for each fl/fl and Δ/Δ group were injected, doses ranging from 1000 to 50 000, into sub-lethally irradiated mice (total of 35 mice per group). (B) Expansion of MA9 GFP+ cells in peripheral blood of mice transplanted with 50 000 fl/fl or Δ/Δ leukemic cells (n = 7/group). (C) Expansion of MA9 GFP+ cells in peripheral blood of mice transplanted with 1000 fl/fl or Δ/Δ leukemic cells (n = 7/group). (D) Overall survival of mice transplanted with limiting doses of fl/fl or Δ/Δ leukemic bone marrow cells (n = 7/cell dose). (E) Analysis using extreme limiting dilution analysis (ELDA) to determine the frequency of leukemia-initiating cells.
Loss-of-KLF4 Impairs Clonogenicity and In Vivo Expansion of Murine L-GMP and Human AML PDX Cells
Previous work confirmed that AML LSCs derived from the MLL-AF9 retrovirus are enriched in the leukemic granulocyte-macrophage progenitor (L-GMP) cell population.48 Immunophenotypic analysis revealed no significant differences in the frequency of GFP+ Sca-1− c-kit+ CD16/32+ CD34+ cells (L-GMP) or other potential immunophenotypically defined LSC populations within the bone marrow of fl/fl and Δ/Δ MA9 mice (Supplementary Fig. S4A–S4C). To evaluate the functional impact of Klf4 deletion in LSCs, we sorted L-GMP cells from fl/fl and Δ/Δ leukemic mice transplanted with cryopreserved primary leukemic cells (Fig. 4A). Serial replating of L-GMP cells in methylcellulose revealed a decrease in the colony-forming capacity of Δ/Δ L-GMP cells (Fig. 4B). Interestingly, this defect was not explained by increased cell death or differentiation (Supplementary Fig. S5A-S5C). However, freshly isolated Δ/Δ L-GMP cells displayed a significant reduction of cells in the G2/M phase of the cell cycle, which was also observed after 48-h culture cytokine-supplemented medium (Fig. 4C, Supplementary Fig. S5D), indicating deregulated cell cycle progression in these LSCs.
Figure 4.
KLF4 knockout murine L-GMPs and human PDX cells show lower clonogenicity and impaired in vivo expansion. (A) Gating strategy to purify L-GMP cells from leukemic bone marrow by cell sorting. (B) Serial colony-forming cell assay in fl/fl or Δ/Δ MA9 L-GMP cells (n = 3/group, mean ± SD). (C) Freshly isolated (0 h) or in vitro cultured (48 h) fl/fl and Δ/Δ L-GMP cells were stained with propidium iodide for DNA content and cell cycle analysis (n = 3/group, mean ± SD). (D) Diagram of KLF4 gene deletion and transplantation in patient-derived xenograft (PDX) cells from an AML patient containing the MLL-AF9 rearrangement (Supplementary Fig. S5). (E) Genome editing of MLL-AF9 PDX cells with Cas9 only (control) or Cas9 and KLF4 sgRNA (KLF4 KO). Edited cells were transplanted into NSGS mice (n = 10/group) and monitored for the expansion of human CD45 leukemic cells in the blood. (F) Kaplan–Meier analysis of survival of mice transplanted with edited PDX cells (n = 10/group). *P < .05, ***P < .001. Two-tailed Student’s t test was used in B, C, and E. Log-rank test was used in F.
To assess whether findings in the syngeneic retroviral MA9-driven model are also relevant in human AML, we developed a patient-derived xenograft (PDX) line from the peripheral blood of a 64-year-old female patient harboring the t(9;11)(p22;q23) translocation (MLL-AF9) (Fig. 4D). PDX cells were CD33+ with a small CD16+ subset in the spleen, demonstrating a myeloid leukemia phenotype with some monocytic differentiation (Supplementary Fig. S6A). Human leukemic cells (hCD45+) were purified from transplanted NSG-SGM3 (NSGS) mice, electroporated for delivery of either Cas9 alone or Cas9 with multi-plex sgRNAs targeting the KLF4 gene, and then edited PDX cells were transplanted into NSGS mice (Fig. 4D). Although both Cas9 control and Cas9/sgKLF4 PDX cells showed an expansion of human blast cells in peripheral blood (Supplementary Fig. S6B), mice receiving edited Cas9/sgKLF4 PDX cells displayed slower proliferation of hCD45 positive cells (Fig. 4E) and significantly improved survival (Fig. 4F). Complete blood cell count demonstrated reduced white blood cell count with fewer neutrophils, monocytes, and eosinophils in the blood of animals receiving Cas9/sgKLF4 PDX cells (Supplementary Fig. S6C). To confirm KLF4 editing in PDX cells, we extracted genomic DNA from the peripheral blood and sequenced the region of the human KLF4 gene targeted by our sgRNAs. Disrupted sequence reads were observed after the cut sites were targeted by sgRNAs (Supplementary Fig. S6D). We performed Inference of CRISPR Edits (ICE) analysis (Synthego) to determine the knockout score and detected gene-editing present in each mouse that had received multi-plex sgRNAs to varying degrees (Supplementary Fig. S6E). DNA sequencing of peripheral blood cells collected 20 days apart indicated that KLF4 knockout cells were either steadily maintained or lost over time, perhaps due to competition with non-edited PDX cells (Supplementary Fig. S6F). Altogether, these data suggest that deletion of KLF4 in human MLL-AF9 AML cells impairs expansion and improves overall survival, similar to Δ/Δ MA9 mice.
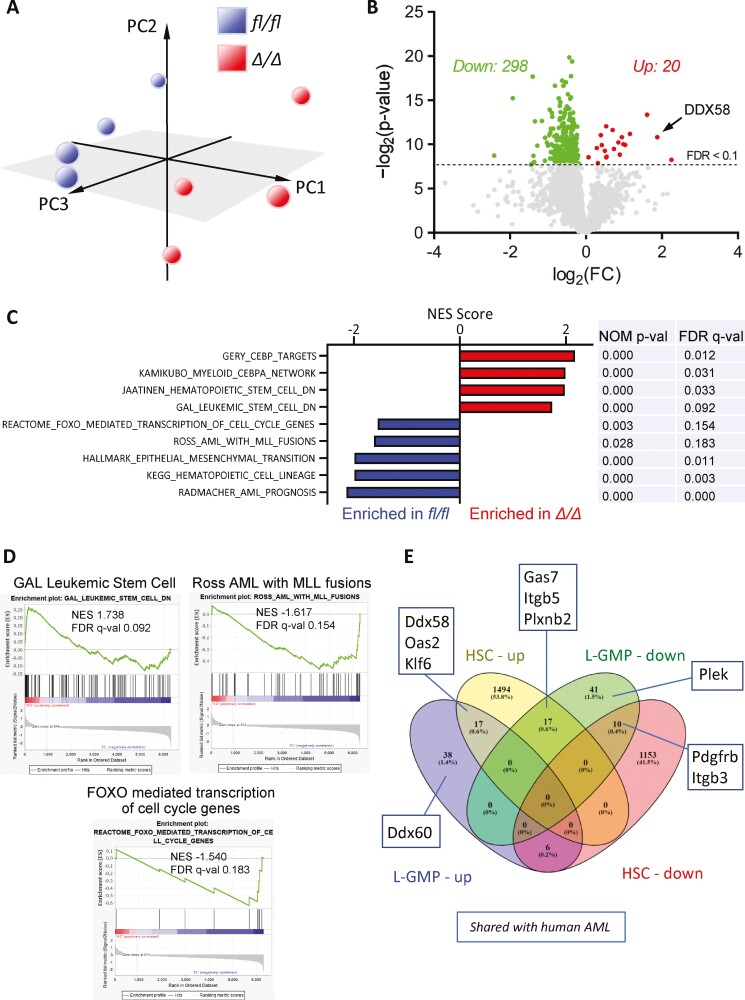
KLF4 Maintains a Leukemic Gene Profile in AML LSCs
To better understand KLF4-dependent changes in the transcriptome of MA9 LSCs, we performed RNA-seq using L-GMPs purified from 4 fl/fl and 4 Δ/Δ leukemic mice as described in Fig. 4A. The principal component analysis showed clustering of fl/fl and Δ/Δ L-GMPs (Fig. 5A). Analysis of individual genes via 2-way heteroscedastic t-test and FDR adjustment (Benjamini-Hochberg correction) revealed 298 genes significantly downregulated and 20 genes upregulated (Fig. 5B, Supplementary Table S1). A gene set enrichment analysis (GSEA) revealed that the transcriptome of Δ/Δ L-GMP cells negatively correlated with gene sets enriched in LSC, MLL-rearranged leukemia, and transcription of cell cycle regulators (Fig. 5C, 5D). Loss-of- KLF4 was also associated with decreased AML and MLL target gene expression (Fig. 5C, 5D). Genes found in AML and MLL fusion sets were found downregulated in Δ/Δ L-GMP cells by qPCR (Supplementary Fig. S7). Loss of EMT signature was also associated with Klf4 deletion, which has previously been reported to drive more aggressive disease phenotypes.49 When we compared genes deregulated by Klf4 deletion in murine HSCs against MA9 LSCs (Fig. 5E), upregulated transcripts associated with inflammatory processes were shared in both Δ/Δ HSC and Δ/Δ L-GMP cells (eg, Ddx58). Genes differentially regulated in the murine model were compared with differentially expressed genes in human AML GMP cells versus normal GMP cells (analyzed from GSE35008) (Fig. 5E), suggesting an overlap between mouse model and human disease. In summary, Δ/Δ MA9 L-GMP cells downregulate the expression of genes associated with MLL-rearranged leukemia and leukemic stemness gene signature.
Figure 5.
Loss-of-KLF4 results in loss of MLL targets and LSC gene signatures. (A) Principal component analysis of RNAseq performed in fl/fl and Δ/Δ L-GMP cells purified from 4 independent leukemic mice per group. (B) Volcano plot of deregulated genes in Δ/Δ L-GMP cells compared to fl/fl L-GMP cells (FDR<0.1). (C) Gene sets significantly enriched in fl/fl and Δ/Δ L-GMP with associated statistical values. (D) Selected GSEA enrichment plots. (E) Peacock plot comparing differentially expressed genes in fl/fl and Δ/Δ normal HSCs and L-GMP cells. Deregulated genes shared between human AML GMP cells and murine MA9 deregulated are indicated in linked boxes.
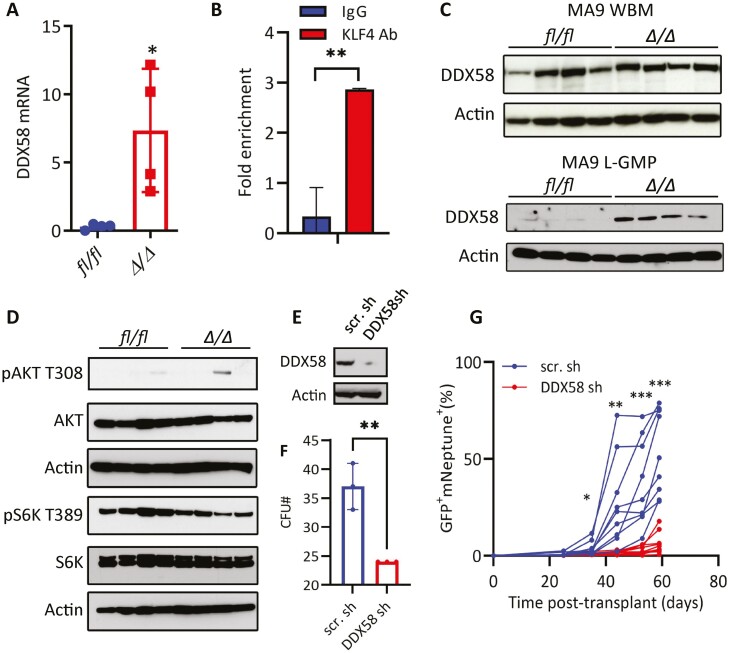
KLF4 Represses the DDX58 Gene in MA9 LSCs
Upregulation of DDX58 in Δ/Δ MA9 L-GMP cells suggested a potential role in MA9 AML. DDX58 (also known as RIG-I) is a dsRNA helicase that acts as an intracellular sensor of viral and endogenous RNAs that activates inflammatory signaling (type I interferon) and suppresses the AKT signaling.50,51 In addition, DDX58 has previously been associated with the regulation of LSCs in a PML-RARα-driven model52-54 and cell cycle regulation in other cancer contexts.55,56 Interestingly, DDX58 was described as a potential transcriptional target of KLF4 according to the CHEA database available through the Harmonizome resource,57 and KLF4 was found enriched by ChIP-Seq in a DDX58 gene regulatory region in murine T-ALL cells which included the presence of a KLF4-consensus binding site.31 We first confirmed the upregulation of DDX58 transcripts in Δ/Δ L-GMPs by qPCR and detected direct binding of endogenous KLF4 protein to the DDX58 5ʹ-promoter region in MA9 L-GMP cells using ChIP-PCR (Fig. 6A, 6B). Immunoblot analysis further confirmed the upregulation of DDX58 protein in purified Δ/Δ MA9 L-GMP cells but not in leukemic (bulk) bone marrow cells from moribund mice (Fig. 6C).
Figure 6.
KLF4 suppression of DDX58 in MLL-AF9 L-GMP cells. (A) qPCR of DDX58 transcripts in fl/fl and Δ/Δ MA9 L-GMP cells purified from 4 independent leukemic mice per group. (B) Binding of endogenous KLF4 to the DDX58 promoter by ChIP-PCR. (C) Immunoblot detection of DDX58 protein in whole bone marrow (WBM) and L-GMP cells from fl/fl and Δ/Δ mice (n = 4/group). (D) Immunoblot analysis of the AKT-mTOR pathway in fl/fl and Δ/Δ L-GMP cells (n = 4/group). (E) Immunoblot of DDX58 in Δ/Δ MA9 scrambled shRNA (scr sh) and DDX58 shRNA (DDX58 sh) Δ/Δ MA9 cells. (F) The colony-forming ability of Δ/Δ MA9 scrambled shRNA (scr.sh) and DDX58 shRNA (DDX58 sh) cells in methylcellulose (n = 3). (G) Expansion of MA9 (Neptune+) and DDX58-shRNA (GFP+) or scramble-shRNA (GFP+) in peripheral blood post-transplantation (n = 10/group). *P < .05, *P < .01, ***P < .001. Two-tailed Student’s t test was used in A, B, F, and G.
To investigate potential mechanisms in AML LSCs, we first investigated whether DDX58 regulates L-GMPs through repression of the AKT pathway because it had been previously reported in LSCs in another AML context.52 Although phosphorylated AKT was barely detectable in purified L-GMP cells (consistent with previous studies),58 decreased levels of phosphorylated S6K, a downstream target of the AKT/mTOR pathway, was detected in some Δ/Δ MA9 L-GMP samples (Fig. 6D). Finally, we knocked down DDX58 in Δ/Δ MA9 cells using lentiviral shRNAs to test whether DDX58 contributes to the regulation of Δ/Δ LSCs (Fig. 6E). DDX58 knockdown significantly reduced clonogenicity in methylcellulose (Fig. 6F). Co-transduction of Δ/Δ LSK cells with retrovirus carrying MA9-mNeptune and DDX58-shRNA-GFP lentivirus and transplantation unexpectedly delayed expansion of GFP+ mNeptune+ cells in peripheral blood (Fig. 6G), suggesting that even though KLF4 regulates DDX58 expression in L-GMP cells, the elevated DDX58 levels do not contribute to impaired LSC frequency. Future studies will be taken to evaluate the role of DDX58 in AML leukemogenesis.
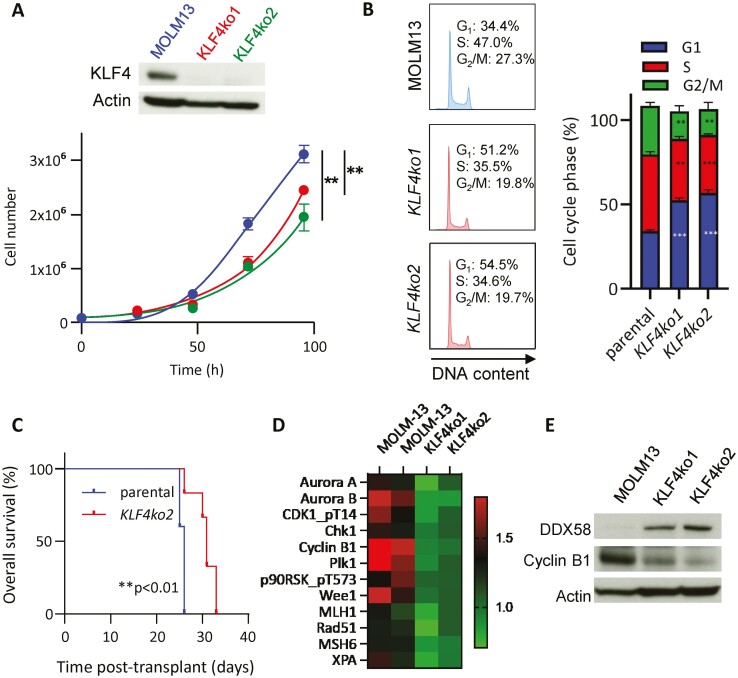
Genome Editing of KLF4 Reduced Expansion and Cycling in MOLM13 Cells
To confirm perturbations induced by loss-of-KLF4 in a human AML cell line with the MLL-AF9 translocation, we deleted the KLF4 gene in MOLM13 cells using CRISPR/Cas9 technology. Two KLF4ko clones were selected from single-cell cloning after genome editing, validated by DNA sequencing, and confirmed KLF4 deletion by immunoblots (Fig. 7A). Consistent with the observations in murine L-GMP cells, both MOLM13 cell clones demonstrated a reduced expansion in culture (Fig. 7A) and a lower percentage of cells in the G2/M phase of the cell cycle (Fig. 7B). Transplantation of KLF4ko2 into NSG mice resulted in prolonged survival compared to the parental line (Fig. 7C). Proteomic analysis of parental and KLF4ko clones by reverse-phase protein array (RPPA) revealed a marked downregulation of proteins involved in the G2/M checkpoint and DNA damage (Fig. 7D), correlating with observations in murine Δ/Δ L-GMP cells. Immunoblot analysis of 2 KLF4 KO clones also demonstrated that upregulation of DDX58 was associated with lower levels of Cyclin B1 (Fig. 7E). In summary, KLF4 deletion in MOLM13 cells results in reduced cell growth, both in vitro and in vivo, associated with impaired G2/M checkpoint.
Figure 7.
Loss-of-KLF4 in MOLM13 impairs cell growth and G2/M cell cycle progression. (A) Confirmation of KLF4 deletion in MOLM13 and KLF4 KO clones via immunoblot. Cell growth of parental MOLM13 cells and 2 clones of KLF4 KO MOLM13 cells edited by CRISPR/Cas9 (mean ± SD). (B) Representative plots of cell cycle analysis by propidium iodide staining of MOLM13 and KLF4 KO cells following 24-h culture (n = 3). (C) Kaplan–Meier analysis of survival of NSG mice transplanted with MOLM13 and KLF4 KO cells (n = 6/group). (D) Heatmap of differentially expressed proteins in MOLM13 (parental) and 2 KLF4 KO cell clones detected by RPPA. (E) Immunoblot of MOLM13 KLF4 knockout detecting Cyclin B1 and DDX58. **P < .01, ***P < .001. Two-tailed Student’s t test was used in A and B. Log-rank test was used in C.
Discussion
Treatment failure of AML patients has been attributed mainly to the genetic complexity of continuously evolving leukemic clones driving the refractory and relapsed disease. Current efforts to identify molecular targets in AML LSCs are critical for developing targeted therapies. A significant limitation is that mechanisms maintaining the LSC pool are largely unknown. Here, we report that the stemness transcription factor KLF4 has pro-leukemic properties in MLL-AF9 AML by establishing a leukemic transcriptome supportive of LSCs.
MLL-rearranged leukemias have a poor prognosis compared to other AML subtypes and harbor drug-resistant LSCs immunophenotypically identified as stem and progenitor cells and maintained by malignant stemness and self-renewal mechanisms.49,59-63 MLL-rearranged leukemias have been characterized by enhanced expression of HOX family transcription factors, typically expressed in the developing embryo but known to be direct targets of MLL fusion proteins.64,65 In addition, there has been an intense focus on epigenetic mechanisms of enhanced MYC expression, a known driver of many cancers, including AML.18,19 The overlap in MLL-AML and embryonic stemness signaling may help explain the unique ability of the MLL-rearranged AML subset to be reverted to a state of induced pluripotency by the expression of 4 stemness factors, including KLF4.66,67 Although previous studies have described the tumor-suppressive function of KLF4 through induction of differentiation and apoptosis, these studies were based mainly on ectopic KLF4 expression in AML cell lines.22,36,37 In contrast, we found deletion of KLF4 is associated with cell cycle defects resulting in reduced clonogenicity and expansion of murine LSCs and patient AML cells upon transplantation. A pro-leukemic function of KLF4 in AML explains why mutations and copy number alterations of the KLF4 gene are not commonly found in the large clinical datasets collected in AML.68,69
Our group has studied the role of KLF4 in normal hematopoiesis and identified unique mechanisms in T-ALL, CML, and AML.70 In pediatric T-cell acute lymphoblastic leukemia (T-ALL) and using the NOTCH1-induced T-ALL mouse model, we described that KLF4 has tumor suppressor function by repressing the MAP2K7-JNK signaling pathway that drives expansion of LICs.31 In contrast, KLF4 promotes LSC self-renewal by repressing the kinase DYRK2 in chronic myeloid leukemia (CML).33 This work describes a pro-leukemic function by sustaining the LSC pool in MA9-induced AML leukemia. Future studies will define the common and unique regulation of KLF4 in LSCs from different leukemias and the commonality of these mechanisms with normal HSCs.
Large-scale clinical studies have allowed for identifying gene expression profiles across AML subtypes. Overall transcript expression of KLF4 in AML is slightly lower compared to healthy bone marrow; however, levels vary by subtype.37 For example, MLL-rearranged AMLs and other M4 and M5 AMLs express higher levels of KLF4 compared to other subtypes (Supplementary Fig. S1A).37 However, these studies have not comprehensively probed putative LIC populations, which are certain to maintain distinct gene expression profiles from bulk disease. In this work, KLF4 deletion resulted in a significant reduction in the frequency of LSC/LIC, which was associated with diminished expression of MLL targets, LSC gene expression profile, and a deregulated G2/M checkpoint. In addition, we identified upregulation of DDX58 in KLF4-deficient cells that were associated with inhibition of mTOR targets and activation of JNK (not shown). Although DDX58 has been extensively studied as a mediator of antiviral response via induction of IFN signaling,50,71 DDX58 is also involved in regulating cellular pathways such as AKT.51,52,54 However, despite being a target of KLF4 in L-GMP, silencing DDX58 in Δ/Δ MA9 AML indicates that it is not involved in promoting KLF4-mediated LSC expansion.
This work provides evidence that KLF4 supports the in vivo cycling of LSCs and maintains a subset of the canonical MLL targets and genes involved in LSC maintenance. KLF4 represses DDX58 in L-GMP, but DDX58 is not involved in the KLF4 regulation of LSCs. Identifying genes involved in the survival and self-renewal of LSCs driving refractory and relapsed leukemia will support the development of LSC-specific therapy.
Supplementary Material
Acknowledgments
We acknowledge Karen Prince for the preparation of figures. This work was supported by the National Cancer Institute to H.D.L. (RO1 CA207086), the ASH Bridge award to H.D.L., the Baylor College of Medicine Comprehensive Cancer Training Program from the Cancer Prevention and Research Institute of Texas to A.H.L. (RP160283), the Cytometry and Cell Sorting Core at Baylor College of Medicine (P30 AI036211, P30 CA125123, and S10 RR024574) and the Flow Cytometry Core at Texas Children’s Cancer and Hematology Center (S10 OD020066).
Contributor Information
Andrew Henry Lewis, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
Cory Seth Bridges, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
David Neal Moorshead, Graduate School of Biomedical Sciences, Baylor College of Medicine, Houston, TX, USA.
Taylor J Chen, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
Wa Du, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
Barry Zorman, Department of Pediatrics, Baylor College of Medicine, Houston, TX, USA.
Monica Puppi, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
H Daniel Lacorazza, Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX, USA.
Funding
Research funded by the National Institutes of Health, National Cancer Institute (RO1 CA207086), and the American Society of Hematology (Bridge Award).
Conflict of Interest
The authors declare no potential conflicts of interest.
Author Contributions
A.L.: designed and performed experiments, collected and assembled data, performed the statistical analysis, and wrote the manuscript; C.S.B.: performed immunoblot analysis; D.M.: knockout gene in cell lines; T.J.C.: performed homing assay; W.D., B.Z.: performed the bioinformatic analysis; P.V.: supervised bioinformatics analysis; M.P.: generated mouse lines; H.D.L.: designed experiments, financed the project, and approved the final manuscript.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Madan V, Koeffler HP.. Differentiation therapy of myeloid leukemia: four decades of development. Haematologica. 2021;106(1):26-38. 10.3324/haematol.2020.262121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kantarjian H, Kadia T, DiNardo C, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11(2):41. 10.1038/s41408-021-00425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roboz GJ, Guzman M.. Acute myeloid leukemia stem cells: seek and destroy. Expert Rev Hematol. 2009;2(6):663-672. 10.1586/ehm.09.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas D, Majeti R.. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129(12):1577-1585. 10.1182/blood-2016-10-696054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shlush LI, Mitchell A, Heisler L, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547(7661):104-108. 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- 6. Vetrie D, Helgason GV, Copland M.. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. 2020;20(3):158-173. 10.1038/s41568-019-0230-9. [DOI] [PubMed] [Google Scholar]
- 7. Ng SW, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433-437. 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 8. Ruan Y, Kim HN, Ogana H, et al. Wnt signaling in leukemia and its bone marrow microenvironment. Int J Mol Sci. 2020;21(17):6247-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang X, Mak PY, Mu H, et al. Disruption of Wnt/beta-catenin exerts antileukemia activity and synergizes with FLT3 inhibition in FLT3-mutant acute myeloid leukemia. Clin Cancer Res. 2018;24(10):2417-2429. 10.1158/1078-0432.CCR-17-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650-1653. 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan J, Takeuchi M, Negishi M, et al. Bmi1 is essential for leukemic reprogramming of myeloid progenitor cells. Leukemia. 2011;25(8):1335-1343. 10.1038/leu.2011.85. [DOI] [PubMed] [Google Scholar]
- 12. Darwish NH, Sudha T, Godugu K, et al. Acute myeloid leukemia stem cell markers in prognosis and targeted therapy: potential impact of BMI-1, TIM-3 and CLL-1. Oncotarget. 2016;7(36):57811-57820. 10.18632/oncotarget.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye M, Zhang H, Yang H, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 2015;17(5):611-623. 10.1016/j.stem.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seipel K, Marques MT, Bozzini MA, et al. Inactivation of the p53-KLF4-CEBPA axis in acute myeloid leukemia. Clin Cancer Res. 2016;22(3):746-756. [DOI] [PubMed] [Google Scholar]
- 15. Kohlmann A, Schoch C, Dugas M, et al. New insights into MLL gene rearranged acute leukemias using gene expression profiling: shared pathways, lineage commitment, and partner genes. Leukemia. 2005;19(6):953-964. 10.1038/sj.leu.2403746. [DOI] [PubMed] [Google Scholar]
- 16. Cimino G, Rapanotti MC, Elia L, et al. ALL-1 gene rearrangements in acute myeloid leukemia: association with M4-M5 French-American-British classification subtypes and young age. Cancer Res. 1995;55(8):1625-1628. [PubMed] [Google Scholar]
- 17. Yokoyama A. Molecular mechanisms of MLL-associated leukemia. Int J Hematol. 2015;101(4):352-361. 10.1007/s12185-015-1774-4. [DOI] [PubMed] [Google Scholar]
- 18. Call SG, Duren RP, Panigrahi AK, et al. Targeting oncogenic super enhancers in MYC-dependent AML using a small molecule activator of NR4A nuclear receptors. Sci Rep. 2020;10(1):2851. 10.1038/s41598-020-59469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bahr C, von Paleske L, Uslu VV, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553(7689):515-520. 10.1038/nature25193. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors [in eng]. Cell. 2007;131(5):861-872. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21. Evans PM, Chen X, Zhang W, et al. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol. 2010;30(2):372-381. 10.1128/MCB.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y, Chen J, Lu C, et al. HDAC1 and Klf4 interplay critically regulates human myeloid leukemia cell proliferation. Cell Death Dis. 2014;5(10):e1491. 10.1038/cddis.2014.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang R, Han M, Zheng B, et al. Kruppel-like factor 4 interacts with p300 to activate mitofusin 2 gene expression induced by all-trans retinoic acid in VSMCs. Acta Pharmacol Sin. 2010;31(10):1293-1302. 10.1038/aps.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307-319. 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Lu T, Sun G, et al. Targeting of apoptosis gene loci by reprogramming factors leads to selective eradication of leukemia cells. Nat Commun. 2019;10(1):5594. 10.1038/s41467-019-13411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alder JK, GeorgantasRW, 3rd, Hildreth RLet al. . Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo [in Eng]. J Immunol. 2008;180(8):5645-5652. 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park CS, Lee PH, Yamada T, et al. Kruppel-like factor 4 (KLF4) promotes the survival of natural killer cells and maintains the number of conventional dendritic cells in the spleen. J Leukoc Biol. 2012;91(5):739-750. 10.1189/jlb.0811413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamada T, Park CS, Mamonkin M, et al. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10(6):618-626. 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan H, Xie L, Leithauser F, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116(9):1469-1478. 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 30. Filarsky K, Garding A, Becker N, et al. Kruppel-like factor 4 (KLF4) inactivation in chronic lymphocytic leukemia correlates with promoter DNA-methylation and can be reversed by inhibition of NOTCH signaling. Haematologica. 2016;101(6):e249-e253. 10.3324/haematol.2015.138172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen Y, Park CS, Suppipat K, et al. Inactivation of KLF4 promotes T-cell acute lymphoblastic leukemia and activates the MAP2K7 pathway. Leukemia. 2017;31(6):1314-1324. 10.1038/leu.2016.339. [DOI] [PubMed] [Google Scholar]
- 32. Li W, Jiang Z, Li T, et al. Genome-wide analyses identify KLF4 as an important negative regulator in T-cell acute lymphoblastic leukemia through directly inhibiting T-cell associated genes. Mol Cancer. 2015;14:26. 10.1186/s12943-014-0285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park CS, Lewis AH, Chen TJ, et al. A KLF4-DYRK2-mediated pathway regulating self-renewal in CML stem cells. Blood. 2019;134(22):1960-1972. 10.1182/blood.2018875922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo X, Tang Y.. KLF4 translation level is associated with differentiation stage of different pediatric leukemias in both cell lines and primary samples [Research Support, Non-U.S. Gov’t]. Clin Exp Med. 2013;13(2):99-107. 10.1007/s10238-012-0187-4. [DOI] [PubMed] [Google Scholar]
- 35. Faber K, Bullinger L, Ragu C, et al. CDX2-driven leukemogenesis involves KLF4 repression and deregulated PPARgamma signaling [Research Support, Non-U.S. Gov’t] [in Eng]. J Clin Invest. 2013;123(1):299-314. 10.1172/JCI64745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morita K, Masamoto Y, Kataoka K, et al. BAALC potentiates oncogenic ERK pathway through interactions with MEKK1 and KLF4. Leukemia. 2015;29(11):2248-2256. 10.1038/leu.2015.137. [DOI] [PubMed] [Google Scholar]
- 37. Morris VA, Cummings CL, Korb B, et al. Deregulated KLF4 expression in myeloid leukemias alters cell proliferation and differentiation through microRNA and gene targets. Mol Cell Biol. 2016;36(4):559-573. 10.1128/MCB.00712-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noura M, Morita K, Kiyose H, et al. Pivotal role of DPYSL2A in KLF4-mediated monocytic differentiation of acute myeloid leukemia cells. Sci Rep. 2020;10(1):20245. 10.1038/s41598-020-76951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ren M, Qin H, Wu Q, et al. Development of ZMYM2-FGFR1 driven AML in human CD34+ cells in immunocompromised mice. Int J Cancer. 2016;139(4):836-840. 10.1002/ijc.30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis AH, Bridges CS, Punia VS, et al. Kruppel-like factor 4 promotes survival and expansion in acute myeloid leukemia cells. Oncotarget. 2021;12(4):255-267. 10.18632/oncotarget.27878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schoch C, Schnittger S, Klaus M, et al. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102(7):2395-2402. 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 42. Townsend EC, Murakami MA, Christodoulou A, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29(4):574-586. 10.1016/j.ccell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wiener J. Pca3d: three dimensional PCA plots. Available at https://cran.r-project.org/web/packages/pca3d/index.html. Published 2020.
- 44. Team RC. R: a language and environment for statistical computing. Available at https://www.R-project.org/. Published 2020.
- 45. Akbani R, Ng PK, Werner HM, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang Y, Hu T, Wang T, et al. AMP-activated protein kinase links acetyl-CoA homeostasis to BRD4 recruitment in acute myeloid leukemia. Blood. 2019;134(24):2183-2194. 10.1182/blood.2019001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818-822. 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 49. Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30(1):43-58. 10.1016/j.ccell.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 50. Rehwinkel J, Gack MU.. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20(9):537-551. 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu J, He Y, Yan M, et al. Dose dependent activation of retinoic acid-inducible gene-I promotes both proliferation and apoptosis signals in human head and neck squamous cell carcinoma. PLoS One. 2013;8(3):e58273. 10.1371/journal.pone.0058273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li XY, Jiang LJ, Chen L, et al. RIG-I modulates Src-mediated AKT activation to restrain leukemic stemness. Mol Cell. 2014;53(3):407-419. 10.1016/j.molcel.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 53. Xu XX, Wan H, Nie L, et al. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell. 2018;9(3):246-253. 10.1007/s13238-017-0431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen L, Cui YB, Si YL, et al. Lentivirusmediated RIGI knockdown relieves cell proliferation inhibition, cell cycle arrest and apoptosis in ATRA induced NB4 cells via the AKTFOXO3A signaling pathway in vitro. Mol Med Rep. 2017;16(3):2556-2562. 10.3892/mmr.2017.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen J, Harding SM, Natesan R, et al. Cell cycle checkpoints cooperate to suppress DNA- and RNA-associated molecular pattern recognition and anti-tumor immune responses. Cell Rep. 2020;32(9):108080. 10.1016/j.celrep.2020.108080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen L, Feng J, Wu S, et al. Decreased RIG-I expression is associated with poor prognosis and promotes cell invasion in human gastric cancer. Cancer Cell Int. 2018;18:144. 10.1186/s12935-018-0639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). 2016:1–16. 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sykes SM, Lane SW, Bullinger L, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146(5):697-708. 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen X, Burkhardt DB, Hartman AA, et al. MLL-AF9 initiates transformation from fast-proliferating myeloid progenitors. Nat Commun. 2019;10(1):5767. 10.1038/s41467-019-13666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Horton SJ, Jaques J, Woolthuis C, et al. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia. 2013;27(5):1116-1126. 10.1038/leu.2012.343. [DOI] [PubMed] [Google Scholar]
- 61. Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852-860. 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Somervaille TC, Cleary ML.. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257-268. 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 63. Ye H, Adane B, Khan N, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. 2016;19(1):23-37. 10.1016/j.stem.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4(3):197-207. 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 65. So CW, Lin M, Ayton PM, et al. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4(2):99-110. 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 66. Chiew MY, Boo NY, Voon K, et al. Generation of a MLL-AF9-specific stem cell model of acute monocytic leukemia. Leuk Lymphoma. 2017;58(1):162-170. 10.1080/10428194.2016.1180683. [DOI] [PubMed] [Google Scholar]
- 67. Liu Y, Cheng H, Gao S, et al. Reprogramming of MLL-AF9 leukemia cells into pluripotent stem cells. Leukemia. 2014;28(5):1071-1080. 10.1038/leu.2013.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-404. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gao J, Aksoy BA, Dogrusoz Uet al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Park CS, Lewis A, Chen T, et al. Concise review: regulation of self-renewal in normal and malignant hematopoietic stem cells by Kruppel-like factor 4. Stem Cells Transl Med. 2019;8(6):568-574. 10.1002/sctm.18-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zevini A, Olagnier D, Hiscott J.. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38(3):194-205. 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.