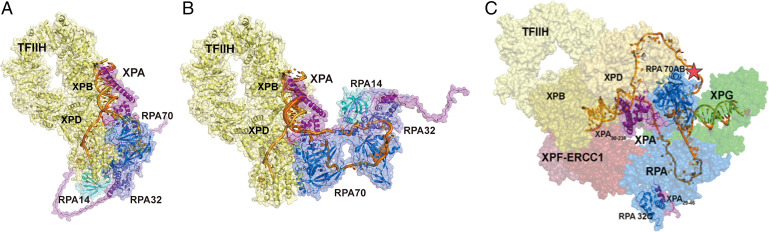
Fig. 7.
Structural models of NER complexes: (A and B) The SAXS models of XPA–RPA–DNA are aligned by superimposing XPA DBD on the XPA–TFIIH cryo-EM structure (PDB ID: 6R04). (A) TFIIH–XPA–RPA bound to a 3′ ssDNA overhang junction. (B) TFIIH–XPA–RPA bound to a 5′ ssDNA overhang junction. (C) Model of the preincision complex bound to the 5′ junction substrate. This model was prepared by aligning XPA DBD in the cryo-EM structure of TFIIH–XPA (TFIIH in yellow) with our XPA–RPA–5′ junction model (XPA in purple and RPA in blue) and placing the endonucleases XPF–ERCC1 (PDB ID: 6SXB) in red and XPG in green (PDB ID: 6Q0W) along the path of the dsDNA. The lesion is indicated as a star on the NER bubble (gold). The two interaction points of XPA and RPA are highlighted as ribbons.