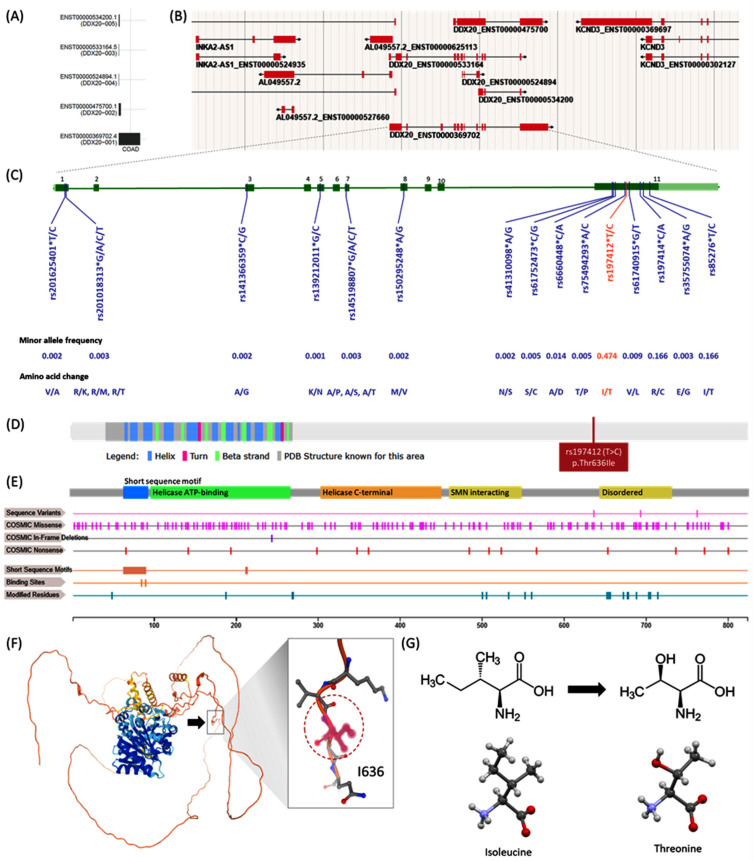
Figure 3.
Structural impact of DDX20 variant. (A) Transcript isoforms in colon adenocarcinoma. ENST00000369702.4 is the most expressed transcript. (B) Genome browser for active DDX20 transcripts in a human female 79 years old. (C) Missense mutations in DDX20 with minor allele frequency >0.001. Amino acid alterations are shown. Nearly nine variants were located within exon 11: c.1907T > C (Substitution, position 1907, T→C), which can lead to amino acid mutation p.I636T (Substitution—Missense, position 636, I→T). (D) Secondary structure of DDX20 protein of 824 amino acids long and molecular weight of 92,241 Da. The location of the studied SNP rs197412 (p.Thr636Ile) is shown. (E) Domains and regions of interest in DDX20 protein (See text). (F) 3D structure of DDX20 protein. The amino acid residue 636 (black arrow) is within an unstructured region in isolation. (G) Functional prediction of rs197412 missense mutation. Chemical structure of Isoleucine and skeletal formula Structure of L-threonine determined by single crystal X-ray diffraction. Color code: Carbon, C: grey; Hydrogen, H: white; Nitrogen, N: blue; Oxygen, O: red. Model manipulated in Avogadro 1.2 and image generated in CCDC Mercury 3.8. Based on the sequence annotation in the neighborhood (phosphoserine of SER-652, 654, and 656 residues) and physicochemical property of the reference and variant residues and reside alteration implicated, the change from medium size and hydrophobic Isoleucine to medium size and polar Threonine was forecasted to be likely benign by functional predicted website tools. Data source: UniProt Q9UHI6 (www.uniprot.org), COSMIC-3D (cancer.sanger.ac.uk/cosmic3d/), AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/Q9UHI6), and Wikipedia (https://en.wikipedia.org/wiki/).