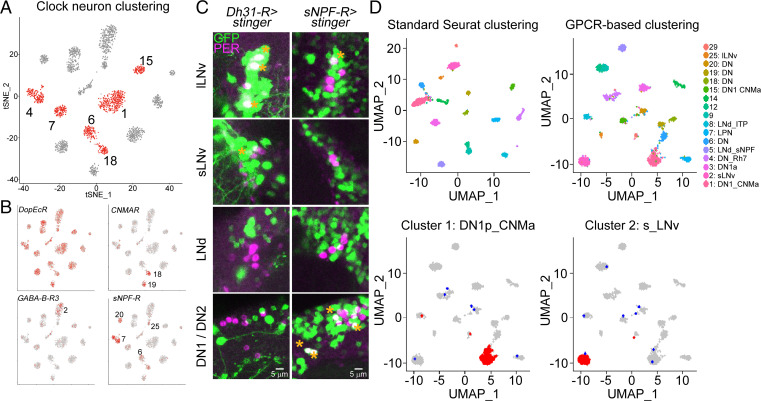
Fig. 2.
GPCRs are differentially expressed and can define clock neuron identity. (A) scRNAseq of clock neurons (clk856 > EGFP) identifies 17 bona-fide clock neuron clusters. DN1 neurons can be separated into six different clusters (1, 4, 6, 7, 15, and 18, labeled in red). For details on clustering see ref. 27. (B) Expression of different GPCRs within these 17 clusters. DopEcR is highly expressed in all clock neuron clusters, whereas CNMaR, GABA-B-R3, and sNPF-R are more differentially expressed. For details see "GPCRs are differentially expressed within the clock neuron network" section. (C) Immunohistochemistry of whole-mount brains stained against GFP (green) and PER (magenta). Nuclear GFP (UAS-stinger) was expressed under the control of GAL4-knock-in lines. Dh31-R-GAL4 drives expression in three out of four lLNvs and one sLNv, and does not express in DN1s, DN2, or LNds. sNPFR-GAL4 is expressed in 1 lLNv, 2 DN1s, and the DN2 neurons. (D) Seurat clustering of individual neurons included in 17 high-confidence clusters. Unsupervised clustering (Top Left) recapitulates previously published results. Similarly, clustering only based on GPCR expression generated 17 different clusters (Top Right). Bottom row: Analysis of retained (red) or newly assigned (blue) clock neuron identities when plotted only based on GPCRs. Most neurons in clusters 1 (Bottom Left) and cluster 2 (Bottom Right) are assigned the same clusters (red dots), whereas only a few cells were assigned to different clusters (blue dots) based on GPCR-only clustering of clock cells.