Fig. 3.
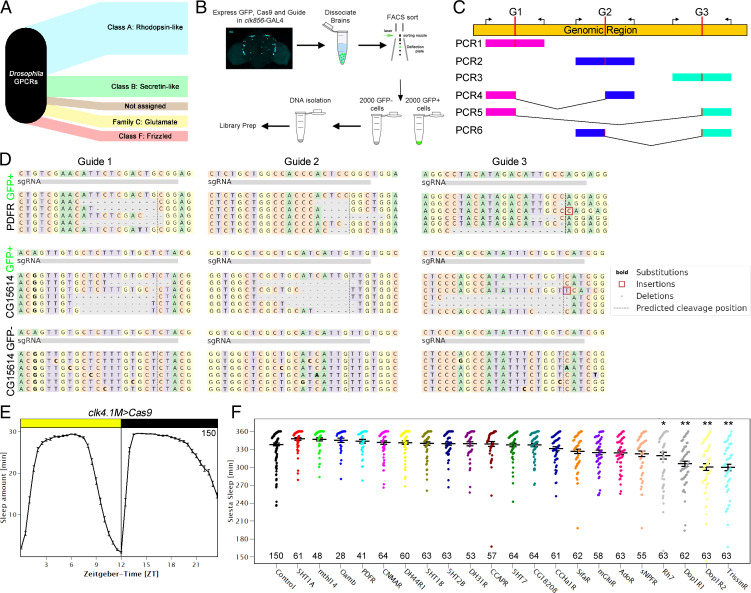
A guide RNA library for all GPCRs of the fly genome allows for cell-specific mutagenesis. (A) Drosophila GPCRs can be categorized into 5 distinct groups (reviewed in ref. 4) (B) Workflow of the targeted genomic sequencing assay. Clock neurons were labeled with EGFP (clk856 > EGFP) while expressing Cas9 and the guide RNA of interest which allows for cell-specific manipulations. GFP-positive cells were analyzed for possible mutagenesis. GFP-negative cells were used as controls for cell specificity. (C) Schematic of genomic sequencing approach. Each genomic region was targeted by three independent guide RNAs (G1 to G3) that led to double-strand breaks (DSBs) at the desired locations (indicated in red). We simultaneously used three sets of primers for each reaction amplifying ∼250 bp. Depending on the timing of guide-RNA-mediated DSBs, we expect either short deletions (PCR1 to 3) or big deletions (PCR4 to 6) in the event of two guides cutting at the same time. (D) Genomic sequencing results of targeted genomic sequencing. Top panel: Sequences of GFP-positive cells of clk856 > EGFP, Cas9, PDFR-g flies. All three guides are able to cause small deletions at the expected site of DSB. Middle panel: Sequences of GFP-positive cells of clk856 > EGFP, Cas9, CG15614-g flies. All three guides are able to cause small deletions at the expected site of DSB. Lower panel: Sequences of GFP-negative cells of clk856 > EGFP, Cas9, CG15614-g flies. No deletions were detected in GFP-negative cells. (E) Sleep behavior of control flies ± SEM expressing Cas9 in DN1 clock neurons (clk4.1M > Cas9). The flies showed the expected sleep pattern with low sleep in the morning and evening and high levels of sleep at night and during the siesta indicating that expressing Cas9 in the dorsal neurons does not affect normal sleep behavior. n = 150. (F) Sleep amount of flies with mutated GPCRs during the siesta (ZT3 to 9) in LD. Clk4.1M > Cas9 flies were used as controls, for experimental flies, Clk4.1M > Cas9 flies were crossed to UAS-guide-RNA lines as indicated on the x-axis. Four guide RNAs significantly reduced sleep during the siesta. Numbers indicate the number of flies used for the behavioral screen. Statistical analysis was performed using a one-way ANOVA followed by a post hoc Tukey pairwise comparison. *P < 0.05 and **P < 0.01 compared to control flies.