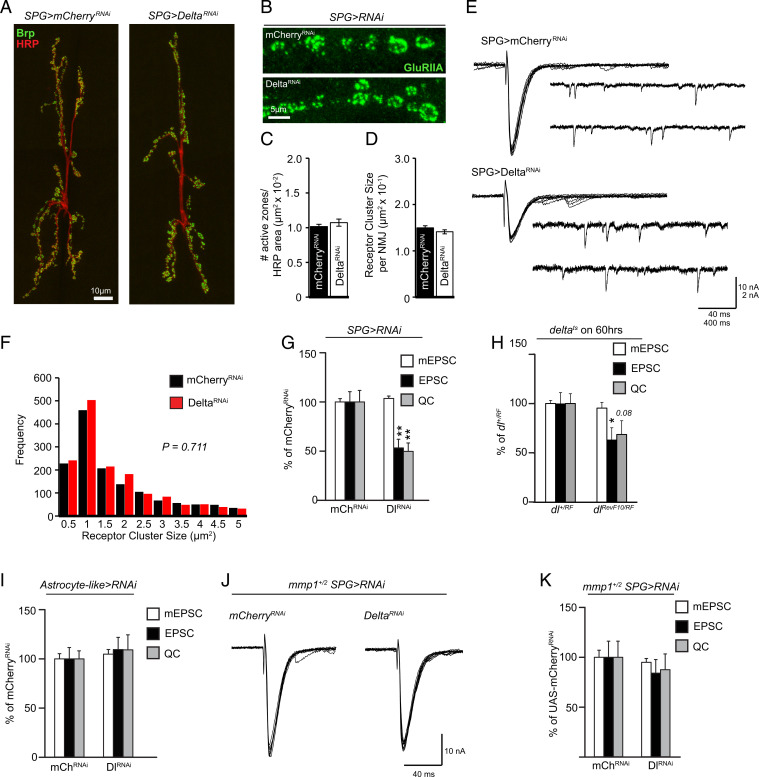
Fig. 4.
Delta regulates synaptic strength at the NMJ. (A and B) Maximum-intensity projections of the NMJ of A3M6/7 of moody-GAL4/UAS-mCherryRNAi or UAS-DeltaRNAi stained with (A) anti-HRP (red) and anti-Brp (green) or (B) anti-GluRIIA (green). Images in A are composites of two high-magnification images. (C) Quantification of the number of active zones defined by anti-Brp puncta in A (n = 18 for each genotype) followed by Student’s t test. (D) Quantification of the average GluRIIA cluster size per NMJ in B (n = 18 for each genotype) followed by Student’s t test. (E) Representative traces of mEPSCs and EPSCs from moody-GAL4/UAS-mCherryRNAi or UAS-DeltaRNAi. (F) Distribution of GluRIIA receptor cluster sizes in D followed by the Kolmogorov–Smirnov test. (G) Quantification of mEPSCs, EPSCs, and quantal content (QC) from E, mCherryRNAi (n = 13) or DeltaRNAi (n = 14), followed by Student’s t test for each respective pair. (H) Quantification of mEPSCs, EPSCs, and QC from delta+/RF (n = 12) or deltaRF/RevF10 (n = 10) followed by Student’s t test for each respective pair. (I) Quantification of mEPSCs, EPSCs, and QC from alrm-GAL4;UAS-mCherryRNAi (n = 14) or UAS-DeltaRNAi (n = 12) followed by Student’s t test for each respective pair. (J) Representative traces of EPSCs from mmp1+/2;moodyGAL4/UAS-mCherryRNAi or UAS-DeltaRNAi. (K) Quantification of mEPSCs, EPSCs, and QC from J, mCherryRNAi (n = 9) or DeltaRNAi (n = 7), followed by Student’s t test for each respective pair. *P < 0.05 and **P < 0.01. All error bars are standard error of the mean.