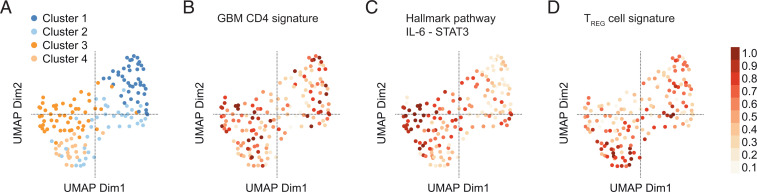
Fig. 7.
GBM transcriptomes of the TCGA dataset are clustered according to TH17 and TREG cell signatures. (A) UMAP representation of the transcriptomes of the TCGA GBM cohort (n = 154). (B–D) GSVA was run on the variance-stabilized expression data of the TCGA cohort for the indicated gene sets. Resulting GSVA scores were normalized to the interval 0 to 1 and projected onto the UMAP representation. (B) Projection of our GBM CD4+ T cell signature (i.e., DEGs in GBM vs. normal-appearing brain CD4+ T cells). (C) Projection of the IL-6–JAK–STAT3 hallmark pathway signature. (D) Projection of a published TREG cell signature (GSE15659). Note that the GBM CD4+ TIL signature coincides with clusters 1 and 3. The IL-6–JAK–STAT3 signature projected on cluster 3, and the TREG cell signature projected on cluster 1 (and 4).