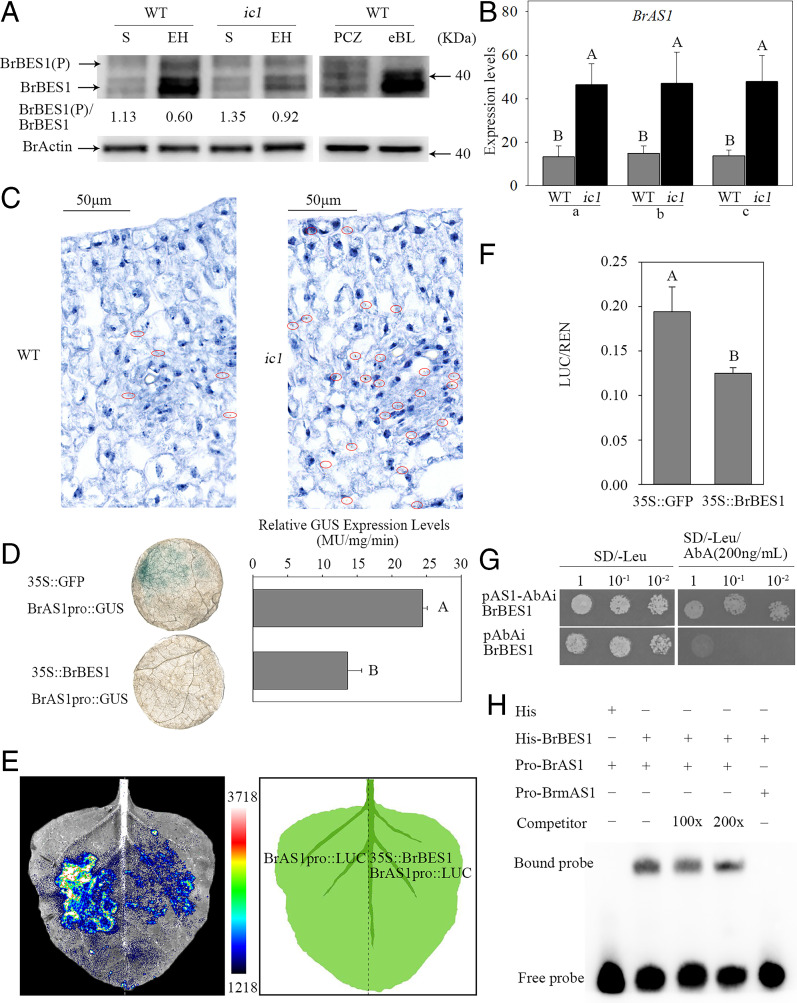
Fig. 5.
BrBIN2 regulates expression patterns of BrAS1 in a BrBES1-dependent manner. (A) The phosphorylation levels of BrBES1 in WT and ic1 plants at the seedling (S) and early heading (EH) stages. The BES1 antibody was used to detect the abundance and phosphorylation of BrBES1 in Chinese cabbage leaves. The actin antibody was used to detect the abundance of BrActin, as a control. The ratio of phosphorylated/dephosphorylated BrBES1 represented average value from three repeats. The repeat included five leaves each from independent plants. (B) Transcript level of BrAS1 in different leaf sections. a, b, and c correspond to the leaf samples shown in SI Appendix, Fig. S7A. (C) The tissue locations of BrAS1 detected via RNAscope ISH at the early heading stage. The brown dots inside the red circles indicate BrAS1. (D) N. benthamiana leaves stained for BrAS1pro::GUS expression upon coexpression with 35S::GFP (upper) or 35S::BrBES1 (lower) and quantification of BrAS1pro::GUS expression in the presence of 35S::GFP or 35S:: BrBES1, respectively. Error bars, SD (n = 12). Significance was determined by ANOVA. (E) The luminescence signal produced by BrAS1pro::LUC in the presence or absence of 35S::BrBES1 in N. benthamiana. (F) Firefly and Renilla luciferases were measured in the presence of 35S::GFP or 35S::BrBES1 using a dual-luciferase reporter system. (G) BrBES1 bound to the BrAS1 promoter in a Y1H assay. (H) BrBES1 bound to the E-box of the BrAS1 promoter in an EMSA.