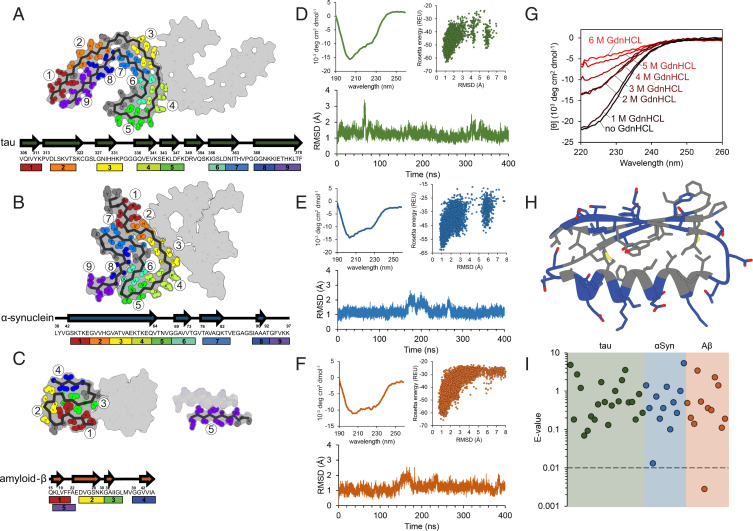
Fig. 2.
Biophysical characterization of designed inhibitors. (A–C) Multiple binding sites on each amyloid fibril structure were selected for targeted inhibitor design. The inhibitor scaffolds were systematically docked to different sites along the fibril ends (each number/color corresponding to a unique binding site). The chosen binding sites correspond to particular β-strand segments (shown in arrows) occurring along the protein chain for tau (A) αSyn (B), and Aβ (C). (D–F) Initial biophysical characterization of each design consisted of CD spectroscopy (Top Left), ab initio structure prediction (Top Right), and long-range molecular dynamics simulations (Bottom). Inhibitors iTau-N (D), i αSyn-F (E), and iAβ-H (F) are shown to maintain stable folds both computationally and experimentally. (G) To assess the stability of the designed inhibitors, CD measurements were taken after a 20-min incubation with increasing concentrations of the denaturant GdnHCl. iTau-N (shown) remains completely stable in 1 M GdnHCl. GdnHCl denaturation curves do not necessarily show a full cooperative unfolding transition, but may indicate destabilization of the folded miniprotein. (H) The fold of each designed inhibitor is driven by a hydrophobic core region (gray residues) surrounded by an exterior of charged and polar residues (blue) (inhibitor iTau-N shown). (I) Each inhibitor was generated de novo, with no apparent homology to known naturally occurring proteins. BLAST E-values, a metric indicating protein homology, demonstrating the designs are well above the significance threshold of 0.01, for all inhibitors targeting tau (green), αSyn (blue), and Aβ (orange), with the exception of iAβ-L.