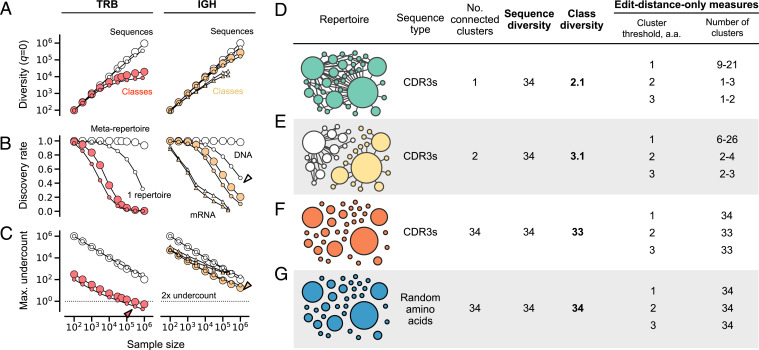
Fig. 3.
Robustness, validity, and comparison to edit distance–only measures. (A) The 0D and 0DS diversity, (B) discovery rate, and (C) maximum error for sequences (open symbols) and classes (filled circles) for repertoires from DNA (small circles) or mRNA (small triangles) and for metarepertoires (large circles) vs. sample size. Maximum undercount in C is the maximum fraction by which sample diversity will underestimate overall diversity (49). Red arrowhead, underestimate for a 300,000-sequence TRB repertoire is ≤33%; yellow arrowhead, sample class diversity of a 1-million-sequence IGH repertoire will underestimate overall class diversity by ≤30×; open arrowhead, for a million-sequence IGH repertoire from DNA, there is a ∼50–50 chance that the next sequence will be new. (D–F) Validity: sequence vs. class diversity for four in silico repertoires, each with 34 unique/752 total sequences with identical sequence frequency distributions (compare Fig. 1B). In the networks, each node represents a unique sequence; node size reflects that sequence’s frequency in the repertoire. Edges connect sequences that differ at a single amino acid position. (D) CDR3s from a somatically hypermutated IGG clonotype. The extent to which class diversity exceeds one reflects intraclone diversity. (E) CDR3s from two different IGG clonotypes. (F) CDR3s drawn randomly from repertoires in this study. (G) Non-CDR3 amino acid sequences generated uniformly at random. Note the contrast between class diversity and edit distance thresholds In D–F, the final two columns, edit distance–based clustering requires a threshold to be chosen: for example, one, two, or three amino acids. Sequences that differ by this threshold amount or less are clustered together. The resulting number of clusters gives one measure of diversity. Different thresholds often give different clusters, and thereby different measures of diversity. In the rightmost column of D–F, note the fairly wide ranges for repertoires A and B, a consequence of the nonuniqueness illustrated in Fig. 2 B–D. In the extremely diverse repertoires in C (all very different CDR3s) and D (random amino acids), edit distance approximates class diversity, but this happens only in the most extreme cases, not in typical repertoires (e.g., Fig. 4 C–E).