Figure 3.
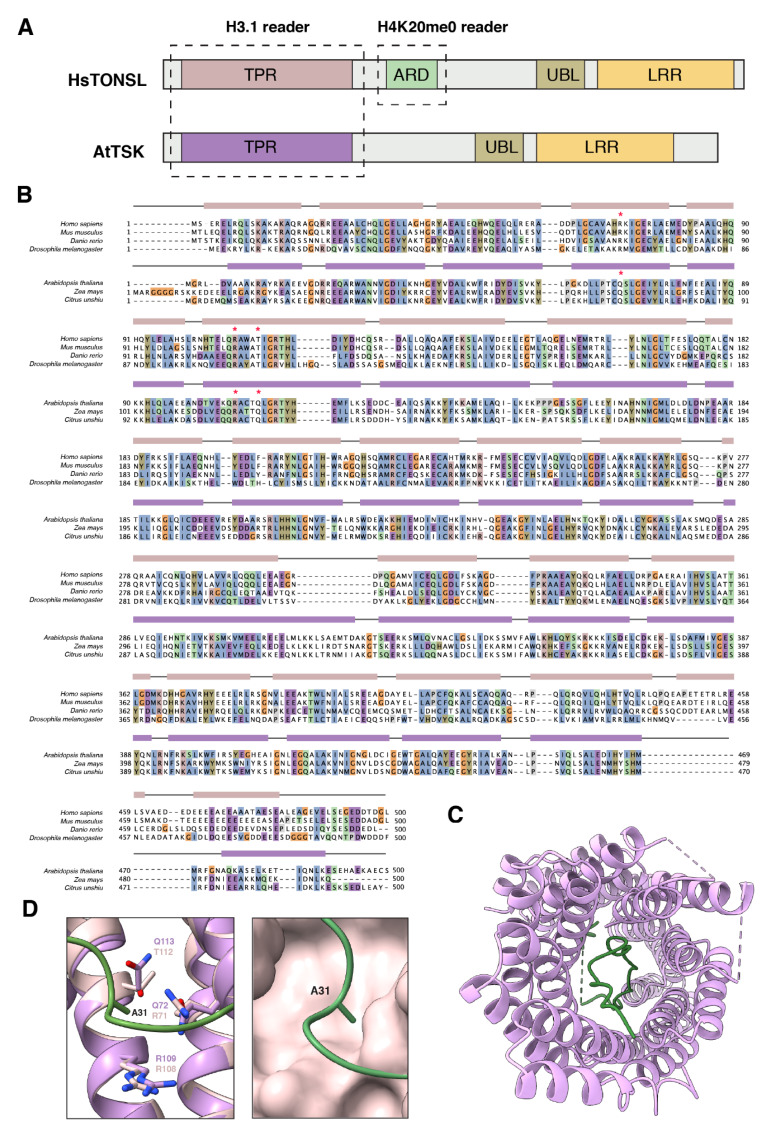
The TPR domain of TSK/TONSL functions as an H3.1 reader: (A) Domain architecture of human and Arabidopsis TONSL/TSK. TPR: tetratricopeptide repeats, ARD: ankyrin repeat domain, UBL: ubiquitin-like, LRR: leucine-rich repeats. The domains in TSK/TONSL responsible for binding specific histones are marked by dashed rectangles; (B) alignment of TPR domains from multiple animal and plant TSK/TONSL orthologs. NCBI reference sequences: NP_038460.4 (Homo sapiens), NP_898914.3 (Mus musculus), NP_001104618.1 (Danio rerio), Q9VSA4.1 (Drosophila melanogaster), NP_188503.2 (Arabidopsis thaliana), PWZ29356.1 (Zea mays), and GAY58445.1 (Citrus unshiu). Residues in the alignment are colored according to the Clustal X color scheme. The α helices of the animal and plant TPR domains of TONSL/TSK, respectively, shown as pink or violet rectangles above the alignment, were predicted by AlphaFold (animals) or based on the crystal structure of the TPR domain of C. unshiu TSK (plants). Red asterisks indicate the residues of the TPR domains shown (plants) or predicted (animals) to mediate specific binding to H3.1 via recognition of residue A31; (C) top view of the TPR domain of TSK (violet) from C. unshiu co-crystalized with the histone H3.1 N-terminal tail (green) (PDB accession number: 7T7T); (D) (Left panel) structural superposition of the solved TPR domain (violet) of plant TSK (C. unshiu) and the AlphaFold-predicted TPR domain (pink) of TONSL (human), with a focus on the histone H3.1A31-binding pocket. The amino acid residues from the TPR domain of plant TSK or animal TONSL interacting with H3.1A31 (green) are shown. (Right panel) Surface representation of the predicted H3.1A31-binding pocket from the TPR domain of human TONSL.
