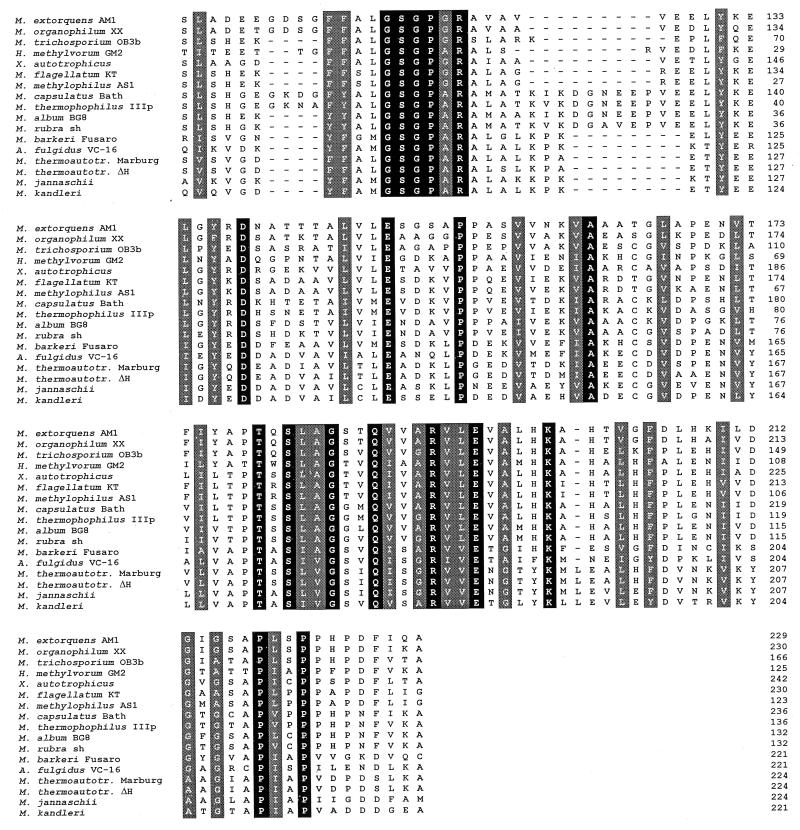
FIG. 1.
Alignment of predicted amino acid sequences corresponding to partial mch genes from bacteria and archaea by the Clustal method (DNAStar). Sequences were obtained by cloning the gene directly from the chromosome from Methylobacterium organophilum XX, Xanthobacter autotrophicus, Methylobacillus flagellatum KT, and Methylococcus capsulatus Bath. Sequences were retrieved by PCR from Hyphomicrobium methylovorum GM2, Methylophilus methylotrophus AS1, Methylococcus thermophilus IIIp, Methylomicrobium album BG8, and Methylomonas rubra 15sh. Sequences from Methylobacterium exotorquens AM1 (11), Methanosarcina barkeri Fusaro (Vaupel and Thauer, EMBL accession no. Y08843), Archaeoglobus fulgidus VC-16 (23), Methanobacterium thermoautotrophicum Marburg (45), Methanobacterium thermoautotrophicum ΔH (41), Methanococcus jannaschii (8), and Methanopyrus kandleri (46) were known before. Amino acid positions conserved in all bacterial and archaeal sequences are shown in black boxes; positions where chemically similar amino acids were found are shown in grey boxes.