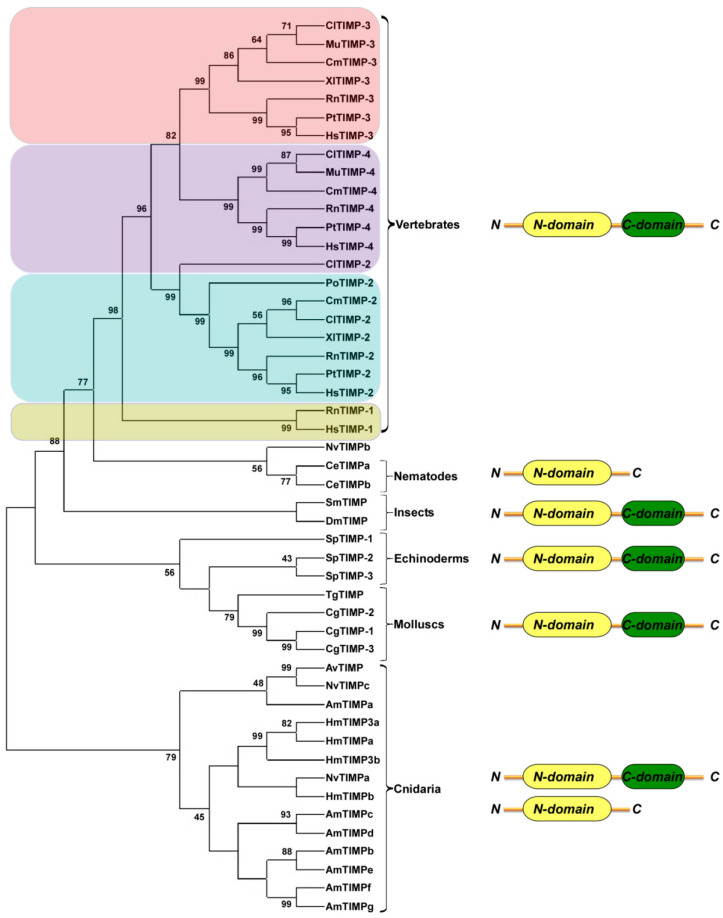
Figure 2.
NJ phylogenetic tree based on the TIMPs from cnidarians to vertebrates. The tree was generated using MEGA X. All the sequences used were obtained from GenBank at National Centre for Biotechnology Information (NCBI). The p-distance model was used to construct the phylogenetic tree. Internal branches were assessed using 1000 bootstrap replications. Bootstrap values greater than 40% are indicated at the nodes. Gene nomenclature is according to [10]. The main taxonomic divisions are indicated. Single- and double-domain TIMPs are shown next to the corresponding groups Different colors represent TIMPs clustering according to isoforms in vertebrates.