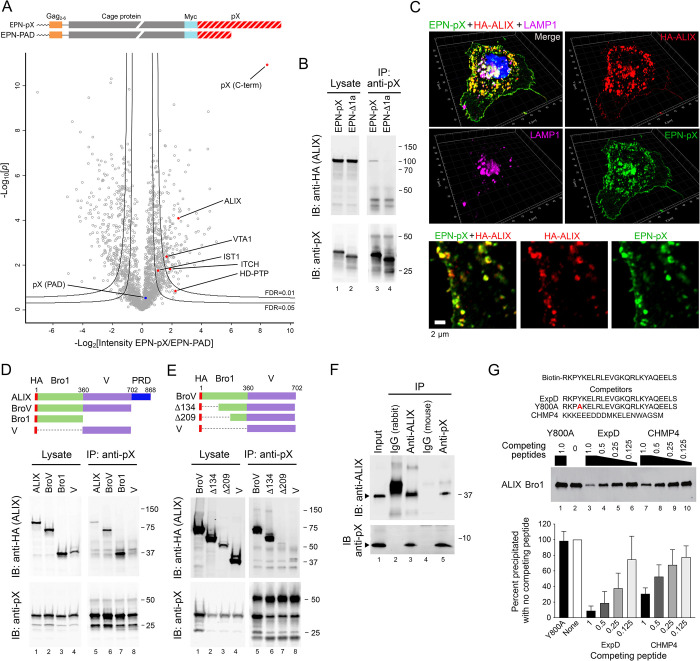
Fig 4. pX interacts directly with the Bro1 domain of ALIX.
(A) EPN-pX constructs expressed in 293T cells for label-free quantitative (LFQ) proteomic analysis of the pX interactome. Below is a volcano plot showing differential abundance of proteins (LFQ intensities) identified in anti-Myc precipitates of EPN-pX versus EPN-PAD lysates. Differences in normalized LFQ peptide intensity are plotted versus significance (p value). Lines represent lower limits of FDR <0.01 and <0.05. Each protein is represented by a single data point with proteins of interest labelled. (B) Co-immunoprecipitation of HA-tagged ALIX with EPN-pX. Proteins in lysates of 293T cells transfected with EPN-pX or EPN-Δ1a were immunoprecipitated with anti-pX, followed by blotting with (top) anti-HA (ALIX) and (bottom) anti-pX. Whole cell lysates are on the left. IP: immunoprecipitation; IB, immunoblot. (C) Merged, single-channel Airyscan fluorescent images of a cell transfected with EPN-pX and HA-ALIX expression vectors, showing pX (green), ALIX (HA, red) and LAMP1 (magenta). At the bottom are shown enlarged dual- and single-channel recordings of the region delineated by the yellow rectangle in the merged image at the top. (D,E) Co-immunoprecipitation of HA-tagged ALIX or the indicated ALIX domain fragments (top) with EPN-pX expressed in 293T cells. (F) Co-immunoprecipitation of bacterially-expressed pX and 6XHis-tagged ALIX Bro1 protein (residues 1–392) mixed in buffer containing 0.7% BSA and 0.07% Tween-20. The input mixture (lane 1) was precipitated with the indicated antibodies, followed by immunoblotting as shown. IgG = isotype control. (G) Biotin-tagged ExpD peptide pulldown assay. Recombinant ALIX Bro1 protein was incubated with synthetic biotin-tagged ExpD peptide (top), affinity isolated on streptavidin beads, and probed with anti-ALIX. Competitor peptides, representing ExpD, the Y800A ExpD mutant, and the C-terminus of CHMP4 [44] (top) were added to the mixture at molar ratios decreasing from 1 to 0.125 relative to the biotin-tagged peptide. At the bottom is shown a quantitative analysis of Bro1 pulled down by biotin-tagged ExpD, normalized to pulldown in the absence of competitor peptide. Results shown are means ± s.e.m. from n = 3 independent experiments.