Figure 3.
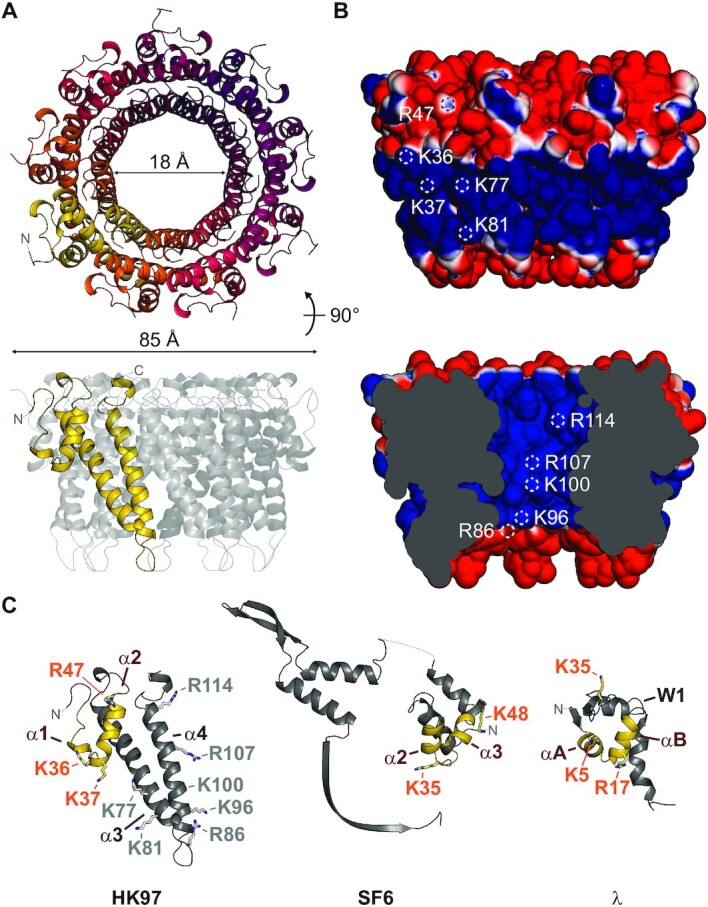
Crystal structure of the HK97 TerS. (A) Ribbon representation of the TerS oligomer, with residues 21–124 displayed. No electron density was observed for residues 1–20 and 125–161. (B) Solvent-accessible surface of the TerS oligomer, colored by electrostatic potential from −1 to 1 kT/e. White circles indicate the location of solvent-accessible positive residues. (C) Comparison of subunit structure with pac phage SF6 GP1 (PDB 3ZQQ) and cos phage lambda gpNu1 (PDB: 1J9I). Putative DNA-binding helix-turn-helix motifs are colored in yellow. Residues important for DNA binding in SF6 and lambda are displayed as sticks. Corresponding residues in the HK97 TerS are indicated with orange text.
