Abstract
The use of multiple drugs simultaneously targeting DNA is a promising strategy in cancer therapy for potentially overcoming single drug resistance. In support of this concept, we report that a combination of actinomycin D (ActD) and echinomycin (Echi), can interact in novel ways with native and mismatched DNA sequences, distinct from the structural effects produced by either drug alone. Changes in the former with GpC and CpG steps separated by a A:G or G:A mismatch or in a native DNA with canonical G:C and C:G base pairs, result in significant asymmetric backbone twists through staggered intercalation and base pair modulations. A wobble or Watson–Crick base pair at the two drug-binding interfaces can result in a single-stranded ‘chair-shaped’ DNA duplex with a straight helical axis. However, a novel sugar-edged hydrogen bonding geometry in the G:A mismatch leads to a ‘curved-shaped’ duplex. Two non-canonical G:C Hoogsteen base pairings produce a sharply kinked duplex in different forms and a four-way junction-like superstructure, respectively. Therefore, single base pair modulations on the two drug-binding interfaces could significantly affect global DNA structure. These structures thus provide a rationale for atypical DNA recognition via multiple DNA intercalators and a structural basis for the drugs’ potential synergetic use.
INTRODUCTION
Small molecules can regulate DNA function to control or treat a variety of human diseases (1,2). These compounds continue to be a focus of drug development programs aimed at creating new generation treatments for cancer (3–5), including as potent payloads for antibody-directed therapy. To date, various types of interactions between small molecules and DNA have been reported, including intercalation, groove binding, covalent binding, and DNA cleavage (6,7). Intercalators have been extensively studied and developed as drugs or diagnostic agents for the treatment of various cancers (8,9). They stack through their planar heterocyclic rings between adjacent base pairs and exert reversible effects on DNA conformation. They exert primary clinical effects by initially inhibiting processes such as DNA replication and transcription or via topoisomerase inhibition, thereby interfering with the function of rapidly proliferating cancer cells (10). The structural basis for the anticancer effects of DNA-targeting compounds is normally studied with one drug at a time. However, it has also been proposed that a combination of two or more DNA intercalators can bind more tightly to specific sites on the DNA, thereby extending sequence specificity (11). Indeed, conjugation of two different DNA-intercalating compounds has been shown to enhance their binding properties, and leads to improved therapeutic potential against some cancers (12–14). Understanding the structural basis of DNA recognition using two or more DNA intercalators may reveal their action mechanism, leading eventually to new therapeutic or diagnostic applications.
Actinomycin D (ActD) and echinomycin (Echi) are two sequence-specific DNA intercalators that share many similar characteristics (Figure 1A, B). Both antibiotics are minor groove intercalators, with ActD recognising 5'-GpC sites and Echi 5'-CpG sites of DNA, with their cyclic peptides stabilising the DNA backbone from the exterior (15–17). Both ActD and Echi can make specific contacts with guanine-NH2 and -N3 atoms. However, the main difference between these two antibiotics is that ActD is a mono-intercalator whereas Echi is a bis-intercalator. ActD has been established in clinical practice for the treatment of cancers such as Wilm's tumor, Ewing's sarcoma, and rhabdomyosarcoma (18,19). Echi has also been considered as a potential clinical drug for the treatment of multiple cancers, although it is highly toxic as a single agent and has yet to demonstrate its clinical efficacy, as revealed by several clinical trials that have not progressed beyond phase II (20,21). Because GC-rich sequences are markers for gene activity regulation, many anticancer drugs have been designed to specifically bind to the GC sites in DNA (22–24). Due to the ability of ActD and Echi to bind to these sequences, these antibiotics thus can serve as paradigms for understanding the principles of drug-DNA intercalation at GC sequences. Furthermore, numerous studies have shown that GC-rich regions tend to mutate, increasing the likelihood of mismatches around GC-rich sequences in the genome (25–27). The occurrence of mismatches can lead to varying degrees of DNA distortion depending on the nature and type of mismatches. For example, purine-purine mismatches such as A:G or G:G have been found to cause greater local structural distortions in DNA (28,29). The purine-related mismatches are also some of the most polymorphic mismatches, and are known to exist in different conformations including wobble, bifurcated hydrogen bonds and various pairing conformations involving anti–syn and anti–anti isomerization (30,31). In addition to mismatches, the formation of Hoogsteen base pairs in DNA can also lead to a local tightening of the diameter of the DNA helix and kinking of the duplex in the major groove (32). The structural differences between these non-canonical base pairs and their Watson–Crick counterparts suggest the importance of Hoogsteen base pairs as potential targets. Therefore, small molecules capable of binding these unusual base pairs in DNA are important tools for therapeutic and fundamental research (33,34). Understanding the structures of such small molecules/DNA complexes can help uncover their specific binding mechanisms, opening new avenues for developing base-pair selective ligands to treat various diseases, including those 20% of human cancers that involve aberrant DNA mismatch repair. Also if the combinations result in novel structural distortions and in synergistic effects in cells and in animal models, these may be able to overcome the resistance produced by either drug alone.
Figure 1.
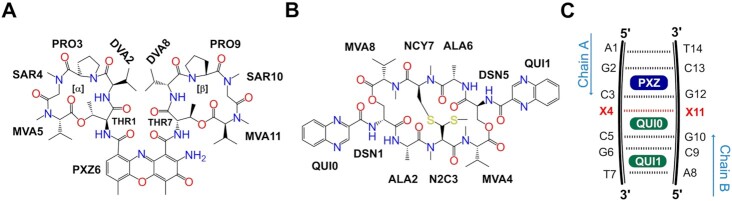
Chemical structures of (A) actinomycin D (ActD) and (B) echinomycin (Echi). The chromophore phenoxazone (PXZ) and two cyclic peptides [α] and [β] in the structure of ActD and the two quinoxaline moieties (QUI) of Echi are shown. The other abbreviations shown in the figure represent l-threonine (THR), d-valine (DVA), l-proline (PRO), sarcosine (SAR), N-methyl-l-valine (MVA), d-serine (DSN), l-alanine (ALA), N-dimethyl-l-cysteine (N2C) and N-methyl-l-cysteine (NCY). The numbers indicate the order of the cyclic peptides in ActD and Echi structures. (C) Schematic representation of ActD-Echi complexed with various mismatch-containing duplexes. Base numbering in chain A and complementary chain B is maintained throughout the study. Central X4:X11 (highlighted in bold red) represents mismatches or Watson–Crick base pair that occurred in this study. PXZ (dark blue) represents the phenoxazone ring of ActD intercalated between the G2pC3/G12pC13 steps; QUI0 and QUI1 (green) are the two quinoxaline rings of Echi intercalated between the C5pG6/C9pG10 steps.
In the current study, we have used a combination of two well-established DNA intercalators, ActD and Echi, and determined the X-ray crystal structures of combination complexes with purine-purine (A:G and G:A) mismatched duplexes, and Watson–Crick (G:C and C:G) base pair-containing DNA duplexes (Figure 1C). Simultaneous intercalation of ActD and Echi results in significant asymmetric distortion in these DNA duplexes, with in each case different geometries at the central base pairs, including wobble-type Aanti:Ganti and Gsyn:Aanti mismatches, two distinct Hoogsteen-type geometries of Gsyn:Canti, and a typical Watson–Crick type of Canti:Ganti base pair. We have also discovered an unusual sugar-edge-mediated geometry in a G‘syn-like’-anti:Aanti mismatch-containing complex, the existence of which has been previously suggested but not observed (35). Depending on the type of central base pair and the nature of the interactions, ActD and Echi were found to cause different degrees of staggering and consequently altered backbone conformation twists. Our results show that swapping of a single base pair at the two drug binding interfaces leads to base pair modulation, which in turn significantly contributes to the formation of different DNA conformers, including features such as single-stranded chair shape, smooth bends in curved shapes, sharp ‘zig-zag’ kink, and a discontinuous four-way-junction like superstructure. These unprecedented polymorphic structural characteristics, along with the base pair modulations proposed in this study, can served as unique recognition signatures of small molecules for cancer-specific DNA targeting.
MATERIALS AND METHODS
Chemicals and oligonucleotides
DNA oligonucleotides were synthesized and purified from MD Bio (Taipei, Taiwan) by polyacrylamide gel electrophoresis (PAGE). DNA oligonucleotide concentration was determined according to Beer's law: A = ϵbc (where A is the absorbance at 260 nm, ϵ the molar extinction coefficient, b the cell path length, and c the molar concentration), using a JASCO-v630 UV-visible spectrophotometer (JASCO International, Tokyo, Japan). The software DNA Calculator of Vladimír Čermák from https://molbiotools.com/dnacalculator.html was used to obtain values of ϵ = 260 nm for each oligonucleotide. ActD and Echi were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Drug concentrations were calculated using Beer's law according to the optical density at 440 nm (ϵ440 nm = 24 500 M−1 cm−1) and 325 nm (ϵ325 nm = 11 500 M−1 cm−1) corresponding to the maximum absorbance of ActD and Echi, respectively. Stock solutions for ActD and Echi were prepared in water and DMSO, respectively used for crystallization.
Crystallization of DNA complexes with ActD and Echi
To grow drug-DNA ternary complex crystals, the DNA oligomers were combined and co-crystallized with ActD and Echi at a molar ratio of 1:1:2 using the vapour diffusion sitting drop method. First, 0.125 mM each of single-strand oligonucleotide solutions were heated to 95°C for 5 min, then annealed on ice for 30 min to allow duplex formation, followed by incubating with 0.250 mM Echi at 4°C for 48 h. Then, 0.125 mM ActD was added and the sample was further incubated for another 24 h. Within a week, small, yellow-coloured crystals were obtained in 5 μl drops containing crystallization buffer equilibrated with 500 μl 30% polyethylene glycol (PEG)-200 at 20°C. To obtain A:G and G:A mismatched complex crystals, d(ACGGGCT)/d(AGCACGT) and d(ACGAGCT)/d(AGCGCGT) DNA oligomers were co-crystallized at a DNA:ActD:Echi ratio of 1:1:2 using a method similar to that described above. d(ACGGGCT)/d(AGCCCGT) and d(ACGCGCT)/d(AGCGCGT) DNA oligomers were used to grow the C:G and G:C Watson–Crick complex crystals with ActD and Echi. Detailed crystallization conditions and incubation temperatures are provided in Supplementary Table S1.
X-ray data collection, phasing and structure refinement
The X-ray diffraction data from single crystals were collected in a synchrotron radiation facility at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan. Diffraction data for A:G, G:A and G:C complex crystals were collected at the NSRRC BL15A1 beamline and recorded with a Rayonix MX300HE CCD area detector. Data for the C:G complex crystal were collected on beamline NSRRC TPS 05A and recorded with a Rayonix MX300HS CCD area detector. The temperature for data collection for all crystals was 100 K. The HKL-2000 program package was used to integrate and reduce diffraction data (36). The phases of A:G mismatched ActD-Echi complex were determined by using molecular replacement (MR) of phaser-MR in a Python-based hierarchical environment for integrated xtallography (PHENIX) v1.10.1 using the partial structure of Echi-d(ACGTCGT)2 (PDB ID: 5YTZ), ActD-d(ATGCTGCAT)2 (PDB ID: 1MNV) and ActD-Echi-d(AGCTCGT)2 (PDB ID: 7DQ0) complexes as templates (37). Initial model building was performed using the crystallographic object-oriented toolkit (COOT) v0.8.9.2 based on the electron density map (38). Structural refinements were performed using the phenix.refine protocol in the PHENIX package. The final refined A:G mismatched complex was used to determine the phases of the G:A mismatch and C:G and G:C Watson–Crick DNA-ActD-Echi complexes. The final crystallographic and refinement statistics of the complexes are listed in Supplementary Table S2. The 2Fo – Fc electron density maps were generated using the fast Fourier transform in CCP4i, and graphical representations of the refined structures were drawn using PyMOL v2.2.3. LigPlot+ was used to analyse the van der Waals interactions between DNA and drug molecules (39). DNA parameters were analysed using the online servers Web-3DNA and CURVES+ (40,41). The values of DNA torsion angles, sugar puckers, local base pair and base-pair step parameters for all complex structures determined in this study are given in Supplementary Tables S3-S5.
RESULTS
Purine-purine A:G and G:A mismatches exhibit distinct backbone conformations via base pair modulation upon intercalation of ActD and Echi
Purine-related mismatches are highly polymorphic in nature and can exhibit dramatic effects on local DNA structures (42–44). It has been recently found that in the case of purine-purine mismatches, base pair swapping (e.g. A:G ↔ G:A) leads to different stacking potentials (45). Therefore, this stacking discrepancy may lead to different stabilising effects and structural perturbations when small molecules bind to the purine-purine mismatches. To understand the effect of intercalation of ActD and Echi on purine-purine mismatches, we solved the crystal structures of two DNA duplexes with a central A:G and G:A mismatch complexed with ActD and Echi in space groups P422 and R3 at 1.95 Å and 2.45 Å resolution, respectively. In both complex structures, the simultaneous binding of two different intercalators leads to an asymmetric distortion of the DNA, with the central mismatch pairs adopting different geometries. A single-strand backbone twisting caused by the different staggering of two drugs also contributes to the structural polymorphism when the A:G ↔ G:A mismatch pairs are reversed. Detailed analysis of these complex structures reveals many unprecedented features of drug–DNA recognition, features that to our knowledge have not been previously reported.
In the A:G structure, each asymmetric unit contained a ternary complex of DNA duplex, ActD and Echi with the central A:G mismatch adopting a more favoured anti-anti geometry (Figure 2A). Three Co2+ ions were found to interact with the N7 atom of guanines G6, G10 and G11, which, along with the coordinated water molecules, appear to stabilize the overall structure. One Co2+ ion was found to stabilize two symmetry-related G6 bases near the Echi intercalation site through octahedral coordination between the N7 atoms of the guanine bases and the water molecules (Supplementary Figure S1A). In addition to cobalt-mediated interactions, analysis of the crystal packing also revealed head-to-head stacking between two asymmetric units. The two ActD ends forms an ‘end-to-end stacking’ by forming ‘sticky’ duplex ends, whereas at the Echi end, the terminal T7:A8 base pair adopts a Hoogsteen conformation and forms a pseudo-continuous duplex, a characteristic feature observed for the binding of two Echi molecules between canonical Watson–Crick base pairs (Supplementary Figure S1B). Interestingly, the G:A mismatch structure contains four ActD–Echi DNA complexes (Supplementary Figure S2A). Superimposition of these structures showed that the two complexes are closely similar (average r.m.s.d. of 0.1 Å) (CPX1 and CPX3, CPX2 and CPX4, CPX refers to a complex), while the complexes CPX1 and CPX2 (or CPX3 and CPX4) are significantly different, indicating the presence of two independent duplexes in an asymmetric unit (average r.m.s.d. of 2.0 Å) (Figure 2B). Therefore, we selected two complexes (G:A-CPX1 and G:A-CPX2) for further structural analysis. In addition to these four drug–DNA ternary complexes, the asymmetric unit also contains three Mg2+ ions, two of which stabilize the interface between CPX2 and CPX4 from the major groove side, and one Mg2+ ion mediates the interface between CPX1 and CPX3 through interactions between OP1 of C3 on the minor groove side (Supplementary Figure S2B).
Figure 2.
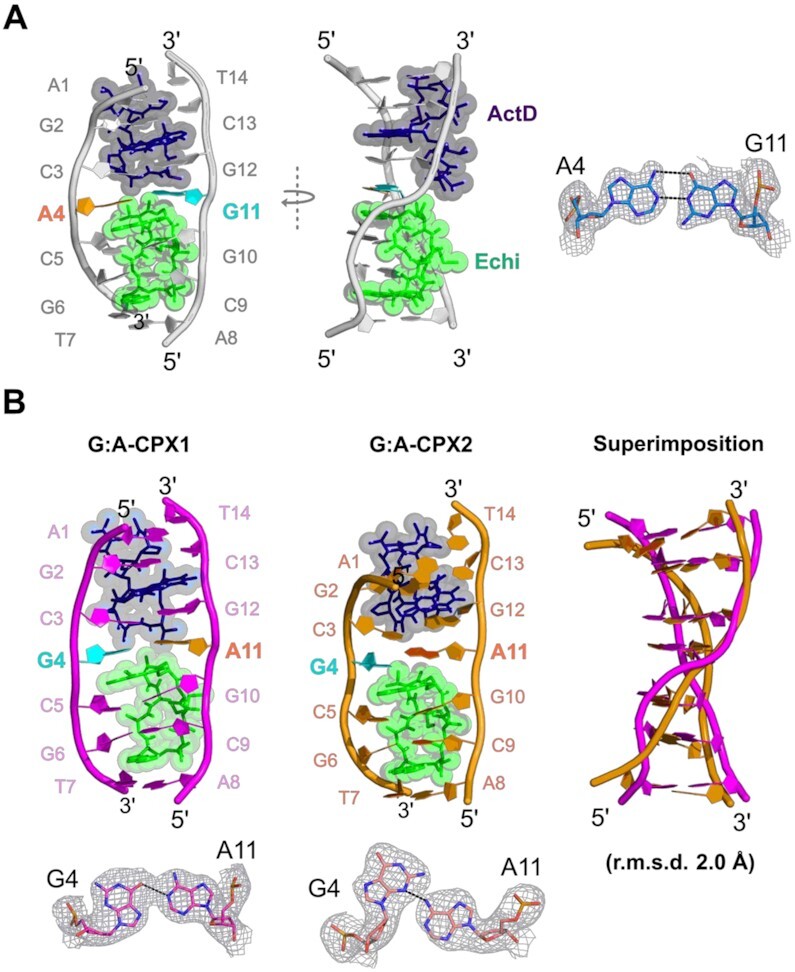
(A) Biological assembly of a crystal structure with a central A:G mismatch in a DNA duplex complexed with ActD and Echi, as shown in front view (left) and side view (right), exhibiting orthogonal asymmetric single-stranded backbone distortion. The DNA backbone and bases are coloured grey. Bases A4 and G11 form a mismatch and are coloured orange and cyan. An enlarged view of the 2Fo– Fc electron density map of the A:G mismatch is shown on the right. (B) The overall crystal structures of two G:A mismatch containing complexes, G:A-CPX1 and G:A-CPX2 with ActD and Echi are shown in pink and orange cartoon representations, respectively. Bases G4 and A11 are highlighted with cyan and orange colours. Superimposition of DNA duplexes in these complexes shows significant differences in the backbone shape with an average r.m.s.d. of 2 Å between the two complexes. An enlarged view of the 2Fo– Fc electron density map of the two distinct G:A mismatches are shown at the bottom.
Intercalation of ActD and Echi at the A:G and G:A mismatch sites results in significant disruption of the DNA backbone geometry compared to typical A or B DNA duplexes. Most nucleotides in the A:G complex adopt C3'- or C4'-exo sugar pucker, whereas in the G:A complexes, the sugar puckers for most nucleotides are in the C3'-endo conformation. The backbone torsion angle γ for most nucleotides at the ActD and Echi intercalation sites remains in the range of 45° to 60°, closer to those of canonical A and B DNA (60° and 40°, respectively), while the central nucleotides X4 and X11 have significant differences in the backbone torsion angles. The γ-values for nucleotides A4, G11 and G4, A11 in A:G and G:A-CPX1 are 171°, 41° and –153°, 10°, respectively, while nucleotides G4 and A11 in G:A-CPX2 have smaller γ-angles of 45° and –84°. The value for torsion angle α for A11 in G:A-CPX2 is 83°, which is significantly different from the values for G11 (–55°) and A11 (-23°) in A:G and G:A-CPX1 pairs, compared to the typical values for canonical A and B DNA (–65° and –41°, respectively). These discrepancies in backbone torsion angles represent distinct conformational features in these duplex structures upon ActD and Echi binding. The DNA helical twist at the ActD-binding interface of the C3pX4 step is lower for A:G and G:A-CPX1 (8° and 21°, respectively), indicating unwinding at this step, whereas the X4pC5 step at the Echi-binding interface showed overwinding, with high twist angle values of 35° and 33° (Supplementary Figure S2C). These alternating twist angles, together with negative roll values, result in a right-handed orthogonal ‘chair’-shaped backbone for one of the DNA strands (chain B) while the other strand (chain A) remains parallel to the helical axis in these two complexes (Figure 3A). On the other hand, the C3pG4 step in G:A-CPX2 has a remarkably high twist angle of about 50°, while the G4pC5 step has a much lower twist angle of about –14°. The 15° positive roll angle of C3pG4 in G:A-CPX2 is accompanied by high twist, leading to bending of the DNA helical axis, resulting in a ‘curved’ duplex in G:A-CPX2. Thus, A:G and G:A-CPX1 exhibit a single-stranded DNA backbone deformation, while G:A-CPX2 has a curved backbone.
Figure 3.
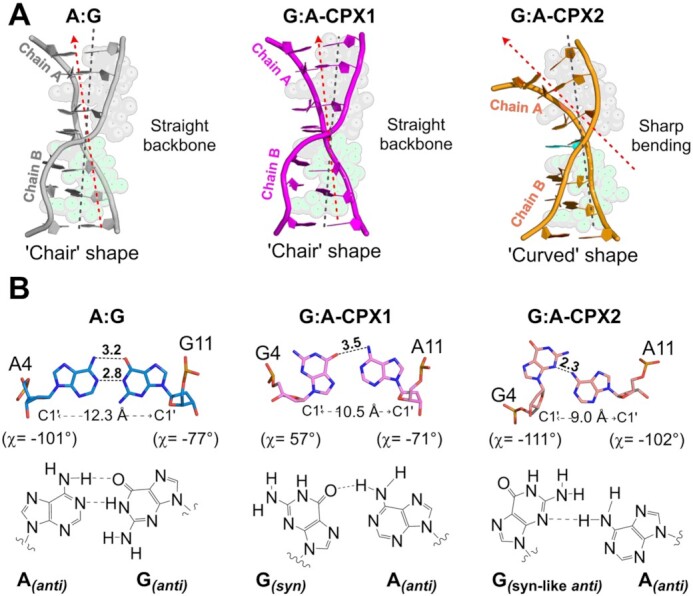
(A) Overall backbone comparison between A:G and G:A mismatch complexes. The A:G complex (grey cartoon) and G:A-CPX1 (pink cartoon) show twisting of only chain B, with the helical axes remaining straight in Chain A. This situation form a ‘chair’ shaped backbone conformation. However, the DNA backbone shows a sharp bend, resulting in a ‘curved’ duplex upon intercalation of ActD and Echi in G:A-CPX2 (orange cartoon). (B) Comparison of central mismatch geometries in A:G and G:A mismatch complexes. The A4:G11 mismatch adopts a common anti–anti geometry with two hydrogen bonds, while the G4:A11 pair in G:A-CPX1 forms a typical syn–anti type wobble base pair with a single hydrogen bond. G4:A11 mismatched pair in G:A-CPX2 forms a sugar-edged ‘syn-like’ anti-anti geometry with a single hydrogen bond. Hydrogen bonds are shown as black dotted lines, and numbers indicate distances in angstroms (Å). Differences in base pair geometries also lead to different C1'–C1' distances at mismatch sites.
In addition to these unique backbone features, we also observed different mismatch geometries for the central A:G and G:A base pairs (Figure 3B). In the A:G complex, the central A4:G11 mismatch adopts an anti-anti conformation, and the glycosidic χ torsion angle has values of –101° and –77° respectively. This type of base pairing has also been observed previously and results in an increased DNA diameter to accommodate the mismatch in the DNA structure (46). The A4 and G11 nucleotides also prefer C3'-exo puckers together with lower base pair shearing in the A:G complex structure. In contrast, the central G4:A11 mispairing in the two G:A complexes adopts more diverse conformations, including syn-anti glycosidic angles (χ ∼ 56° and –71°) in G:A-CPX1 and ‘syn-like’ anti–anti (χ ∼ –111° and –102°) in G:A-CPX2. Previous NMR studies have extensively demonstrated the existence of unusual sheared G:A mismatch geometry in DNA structures (35,47). To our knowledge, this is the first observation that two drugs can capture this unusual G:A mismatch conformation. The G4 nucleotides from both G:A complexes have two different sugar puckers, with a syn conformation preferring C3'-endo pucker, with the ‘syn-like’ anti preferring C3'-exo pucker. The presence of unusual ‘syn-like’ anti-anti geometry generates an extremely high shear value (6 Å) at the G4:A11 pair, causing the G4 base to move into the major groove, while A11 is pushed back into the minor groove (Supplementary Figure S2D). This unusual base pairing further pushes away the N-methyl group of the MVA residue in ActD to avoid the molecular crowding at the A11pG12 step, which results in an slanted intercalation for the ActD chromophore into the G2:C13/C3:G12 step in G:A-CPX2.
Structural details of ActD and Echi binding sites in A:G and G:A mismatch complexes
Figures 4 shows in detail the different stacking and hydrogen bonding interactions between ActD and Echi with DNA in A:G and G:A complexes. In the A:G and G:A complex structures, the phenoxazone ring (PXZ) of ActD is intercalated into the G2:C13/C3:G12 base pair step, while the quinoxaline rings (QUIs) of Echi are bis-intercalated into the X4:X11/C5:G10 and G6:C9/T7:A8 base pair steps. The macrocyclic peptide rings of ActD and Echi stabilize the minor groove by widening the groove and partially unwinding the duplex. The two halves of the A:G and G:A complexes remain asymmetric due to the intercalation of the two different drugs. Intercalation of the PXZ ring of ActD into the A:G and G:A complexes results in extensive stacking with guanine bases on both sides flanking the ring, similar to the previous structures. However, the opposite orientations of the PXZ rings in the A:G and G:A complexes forms different hydrogen-bonding interactions (Figure 4A). In both A:G and G:A complexes, the O1' atom of PXZ form a bifurcated intermolecular hydrogen bond with the O2 and N2 atoms of the G2 and C13 bases, respectively (3.5 and 3.4 Å in A:G, 3.3 and 3.4 Å in G:A-CPX1 and 3.2 Å each in G:A-CPX2). In contrast, the N2 atom of PXZ forms hydrogen bonds with the O4' sugar oxygen of C3 (2.4 Å) in the A:G complex and at distances of 2.9 Å and 2.8 Å of C13 in the G:A-CPX1, respectively, on the other strand. Despite the opposite orientation of the two ActD molecules in these complexes, the more common hydrogen-bonding interactions between ActD and DNA were observed via the CO/NH atom of THR1 and THR7 and the N2/N3 atom of G2 and G12 nucleotides (Figure 4B).
Figure 4.
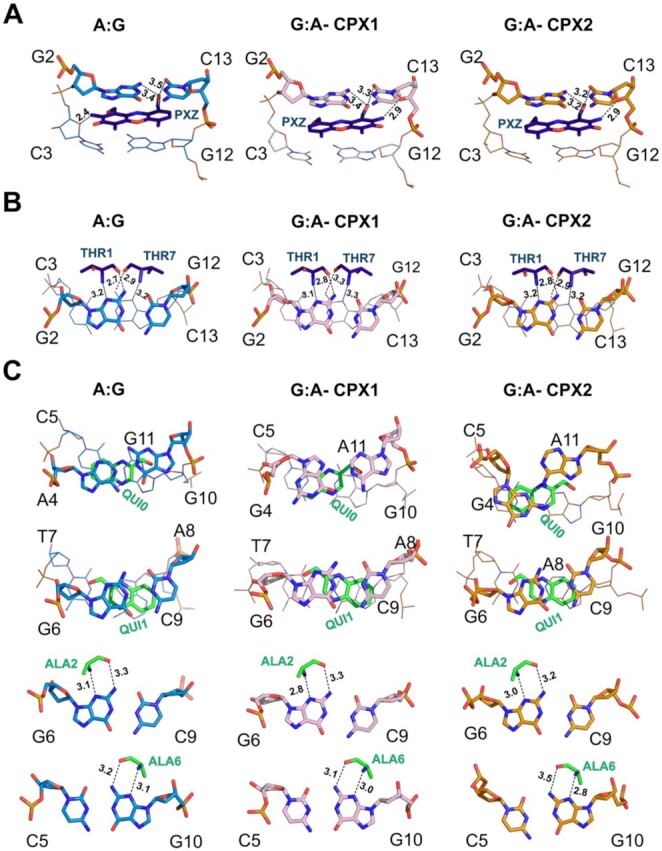
Overall stacking and hydrogen bonding interactions between ActD (dark blue sticks) and Echi (green sticks) and DNA nucleotides in the A:G (light blue sticks), G:A-CPX1 (pink sticks) and G:A-CPX2 (orange sticks) mismatched complex structures. (A) The phenoxazone ring (PXZ) of ActD is intercalated into the G2pC3 step, forming stacking and hydrogen bonding with the bases G2, C13 and C3 in the three complexes. (B) Intermolecular hydrogen bonds to the N2/N3 atoms of bases G2 and G12 and the threonine residue (THR of ActD) in all three complexes are shown. (C) The quinoxaline ring (QUI) and alanine residue (ALA) of Echi are intercalated into the C5pG6 sites, resulting in stacking and hydrogen bonding.
The different stretch distances and opening angles at the central A:G and G:A mismatch sites further cause differences in stacking with the QUI0 ring of Echi (Supplementary Figure S3A). In the A:G complex, the central A4:G11 mismatch has a stretch distance of 1.5 Å and a small opening angle of –2°. The unusual geometries of the two G4:A11 base pairs result in lower stretch distances of 0.4 Å and -1.4 Å, and unusually large base pair openings of 20° and 49° in G:A-CPX1 and G:A-CPX2, respectively. These differences at the binding interface of the two drugs also result in different intermolecular van der Waals interactions with the central mismatch pairs. Ligplot+ analysis shows that the MVA and DSN residues of ActD and QUI0 and MVA residues of Echi consistently form van de Waals contacts with the central mismatches in all three complexes (Supplementary Figure S3B). However, the adoption of a syn geometry by the G4 nucleotide in G:A-CPX1 results in additional interactions with the PRO and SAR moieties of Echi, whereas the ‘syn-like’ anti geometry of G4 in G:A-CPX2 only results in an additional interaction with the PRO residue of Echi. The C5:G10 and G6:C9 base pairs within the quinoxaline intercalation site have alternating negative and positive buckle values, resulting in intermolecular hydrogen bonds between the CO/NH of the two ALA residues and the N2 and N3 atoms of the G6 and G10 bases, which define the pronounced sequence preference of Echi for CpG sites in DNA (Figure 4C).
Canonical C:G and G:C base pair-containing duplexes display considerable base-pairing changes following ActD and Echi intercalation
As described above, base pair swapping in bulky purine-related A:G and G:A mismatches results in distinct backbone conformations and base pair modulation upon intercalation of ActD and Echi. To understand and compare the effects of intercalation of the two drugs on all Watson–Crick base pair-containing duplexes, we have determined two crystal structures of d(ACGGGCT/AGCCCGT) and d(ACGCGCT/AGCGCGT), DNA oligomers with a central canonical C:G and G:C base pair complexed with ActD and Echi in space groups P4222 and P6322 at 1.95 and 2.96 Å resolution, respectively. In contrast to the purine-related A:G and G:A mismatch complexes, the canonical C:G and G:C base pair complexes further contributed to the structural diversity caused by two different intercalators. Although the C:G base pair containing DNA exhibits typical features similar to those of the A:G complex, the base pair swapping from C:G ↔ G:C results in distinct Hoogsteen base pairing along with diverse structural polymorphism. The structural study has shown for the first time that the presence of Hoogsteen base pairing can cause a sharp kink in the DNA backbone and the induction of higher order structural features. The details of the structural features induced by ActD and Echi in canonical C:G and G:C base pair-containing complexes are given below.
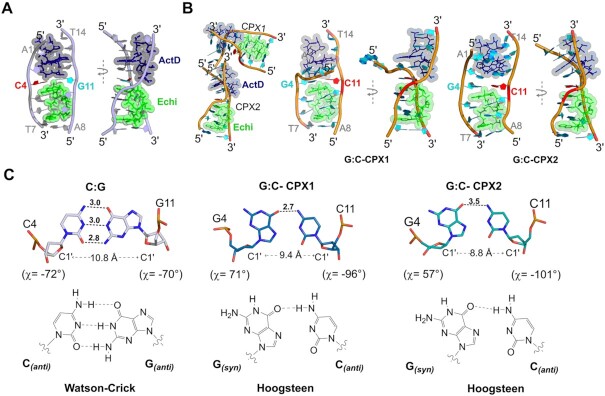
The C:G structure shows a single ternary complex of duplex DNA with ActD and Echi (Figure 5A), with the overall structure further stabilized by end-to-end crystal packing mediated by π-π stacking between the terminal nucleotides of each duplex and lateral side-to-side interactions by the Zn2+ and K+ metal ions and water molecules in the major groove (Supplementary Figure S4). One Zn2+ was found to mediate a symmetry-related interaction with guanine residues. A potassium (K+) ion was also found to mediate the interactions between the terminal phosphate oxygen atoms with two adenine nucleotide A1s in the symmetry-related duplexes. The backbone features of the C:G complex are similar to the earlier A:G mismatch complex, with an asymmetric single-stranded DNA twist. On the other hand, the biological assembly of the G:C structure contains two ternary ActD-Echi DNA complexes, in which a Mn2+ ion interacts with the symmetry-related guanine bases (G6 and G6*) at Echi binding interface through N7-mediated interactions in the major groove (Figure 5B and Supplementary Figure S5).
Figure 5.
Overall structure of C:G and G:C Watson–Crick duplexes in complexes with ActD and Echi. (A) Biological assembly of the crystal structure containing a central C:G Watson–Crick base pair in a DNA duplex complexed with ActD and Echi, as shown in front (left) and side (right) views, with typical single-stranded backbone distortion. The DNA backbone is shown in a white cartoon with the central C4 and G11 bases highlighted in bold red and cyan font. (B) Biological assembly of the crystal structure containing a G:C Watson–Crick base pair shows two separate duplexes after ActD and Echi intercalation, as shown in the orange cartoon. G:C-CPX1 displays backbone with twisted end, while G:C-CPX2 shows a kink in individual DNA backbone strand. The central bases G4 and C11 are highlighted in cyan and red bold. (C) Comparison between the geometries of the central C4:G11 and G4:C11 pairs in the two complex structures. The C4:G11 base pair shows a typical Watson–Crick geometry with both bases in the anti-anti conformations with three hydrogen bonds. The G4:C11 base pair, on the other hand, forms a Hoogsteen base pairing with syn-anti geometries containing two different χ values for syn G4 bases and hydrogen-bonded interactions. The differences in the C1'-C1' distances for each base pair type are shown. Hydrogen bonds are represented by black dotted lines and distances in angstroms (Å).
The insertion of ActD and Echi did result in significant perturbation of the DNA backbone, as shown by their torsion angles. In the C:G complex, most backbone torsion angles δ are in the 130–150° range, closer to typical B-DNA values. Interestingly, nucleotide G4 in G:C-CPX2 has a much lower value, around 87°, which is closer to the value for canonical A-DNA (81°). For the same G4 nucleotide, the backbone torsion angle γ is 177°, significantly higher than the values for C4 and other G4 nucleotides in C:G and G:C-CPX1, respectively. These sudden changes in torsion angles further lead to typical single-strand twisting of the DNA backbone. Interestingly, in G:C-CPX2, the ActD chromophore is intercalated in such a way that the PXZ ring is slanted to the major groove, which causes the C11 nucleotide to be significantly staggered (3.8 Å) into the major groove to avoid wedging into the C11pG12 step via the N-methyl group. The backbone torsion angle γ undergoes a major change from 122° to 44° at the G12pC11 step of chain B, resulting in a sharp kink in the DNA backbone at this step. In G:C-CPX1, the terminal nucleotides A1pG2 in chain A are tilted approximately 90° from the helical trajectory. This striking feature is reflected in a ∼180° change in the β torsion angle (between O5' and C5' of the DNA backbone) of the C3 nucleotide compared to other nucleotides in the same strand. In all three complexes, higher twist angle values at the ActD-binding interface at the C3pX4 step indicate overwinding of the DNA backbone at this step. Interestingly, in the C:G complex, the roll angle at the C3pX4 step is much lower (about 13°) compared to 56° and 46° in the G:C-CPX1 and G:C-CPX2 structures, respectively. These differences are correlated with single-stranded DNA deformation in the C:G Watson–Crick complex, similar to the A:G structure. In addition to differences in twist and roll angles, the C:G and G:C complexes also show significant differences in other DNA parameters such as base pair shift and slide (Supplementary Figure S6). In the C:G complex, the C4pC5 step has a positive shift (0.1 Å) and a positive slide value (2.7 Å) compared to the -4.4 Å and -2.9 Å in shift and -2.8 Å and -4.6 Å in slide for G:C-CPX1 and G:C-CPX 2, respectively. The central C4:G11 base pair in the C:G complex adopts a canonical Watson–Crick anti–anti geometry (χ ∼ –70° and –72°), while the G4:C11 base pairs in the two G:C complexes have two distinct Hoogsteen syn–anti conformations (χ of ∼ 71° and -96° for G:C-CPX1 and ∼ 57° and –101° for G:C-CPX2) (Figure 5C). These syn-anti geometries with two different χ angles result in a narrowing of DNA diameter and reduced C1'-C1' distances compared to the anti-anti conformation, leading to differences in intermolecular interactions in the complexes.
Structural features of ActD and Echi binding sites in C:G and G:C complexes
In the C:G and G:C complexes, the ActD and Echi molecules are intercalated between the base pair steps G2:C13/C3:G12 and C5:G10/G6:C9, respectively, with the cyclic peptides located in the minor groove. Detailed analysis of the intercalation sites of ActD and Echi in these complexes has revealed many known drug-DNA interactions, including stacking, hydrogen bonding, and van der Waals interactions. Extensive stacking interactions with guanine bases flanking the PXZ ring are conserved in both complex structures. However, the ActD PXZ ring shows opposite intercalation orientations in these complexes, with differences in intermolecular hydrogen bonds (Supplementary Figure S7A). In the structure of the C:G complex, the N2 atom of PXZ forms a single hydrogen bond with the sugar oxygen O4' of the C3 base with a distance of 2.7 Å. While in G:C-CPX1 a bifurcated hydrogen-bonded interaction with the sugar oxygen O4' and the backbone oxygen O5' (2.7 Å and 3.2 Å) and a single hydrogen-bonded interaction with the O4' sugar oxygen is formed, 2.8 Å from the C13 base on the other strand. The O1 oxygen atom of the PXZ ring forms bifurcated hydrogen bonding interactions with the N2/O2 atoms of the G2 and C13 nucleotides in all three complexes. Also, the C13 nucleotide in G:C-CPX1 and G:C-CPX2 forms additional hydrogen bonded interactions between the O1' of PXZ and the O2 and O4' oxygen atoms of C13, respectively. The more common hydrogen bonds between the two THR residues of ActD mediated by the CO/NH atom and the N2/N3 atoms of G2 and G12 nucleotides are well preserved in these complex structures (Supplementary Figure S7B). The base pairs C5:G10 and G6:C9 within the quinoxaline intercalation sites show alternating negative and positive buckle parameters, resulting in intermolecular hydrogen bond formation between the CO/NH of the two ALA residues and the N2 and N3 atoms of the G6 and G10 bases, which define the strong sequence preference of Echi for CpG sites of DNA. Interestingly, the –13° opening angle of the G6:C9 base pair, which pulls the guanine G6 towards the major groove, causing an increase in distance between the N3 atom of the guanine and the NH of the ALA3 residue, results in a single hydrogen bond being formed in G:C-CPX1 (Supplementary Figure S7C).
Furthermore, the different geometries of the central base pairs in the C:G and G:C complexes lead to stacking differences with the quinoxaline QUI0 of Echi. The C4:G11 pair in the C:G complex has a very low stretch distance (0.0 Å) and a low opening angle (2°), while the two G4:C11 base pairs in the G:C complex have a higher negative stretch distance (–1.2 and –1.2 Å), with higher base pair opening values (13° and 18°, respectively) (Supplementary Figure S8A). These distinct differences at the central base pair sites result in varying number of drug residues forming van der Waals interactions with the central X4:X11 base pairs in these complexes, as observed with LigPlot+ (Supplementary Figure S8B).
The G:C base pair complex induces a four-way junction-like superstructure at the ActD intercalation site
In the two independent complexes in the asymmetric unit of the G:C crystal structure, the two terminal bases of chain A in CPX1, 5'-A1pG2, are flipped away from the normal helical trajectory at the ActD-binding site, by approximately 90°. These flipped bases exhibit no disorder in electron density maps, confirming the existence of a discrete novel higher order structure (Supplementary Figure S9). Detailed analysis of the symmetry-related complex has revealed a unique superstructure in the form of a discontinuous four-way junction at the ActD-binding site in the G:C complex structure (Figure 6A). This unnatural base flipping causes severe twisting of the DNA backbone, resulting in crossover between the 5'-end of chain A in CPX1 and the symmetry-related 5'-end in CPX1*. The 3'-T14 base in the symmetry related CPX2* chain B forms ‘base-to-base’ π–π stacking with the flipped-out 5'-A1 base of CPX1, resulting in a discontinuous inner strand of a four-way junction superstructure (Figure 6B). The outer strands of the superstructure are composed of stacking interactions between 3'-T14 from Chain B of CPX1 and 5'-A1 residue in chain A of CPX2 (and the symmetry related CPX1* with CPX2*). As a result, the terminus of CPX2 allows the overall structure of the complex to form a complete four-way junction, with two ActD molecules intercalating at the cross-linking interface of the superstructure. Two Echi molecules stabilize the ends of the entire four-way junction structure. When viewed from the top, the two symmetry related complexes CPX1 and CPX1* form an angle to the horizontal plane of the crossover site, where two ActD molecules are intercalated from the minor groove. The crossover junction consists of two ActD PXZs intercalated into G2*:C13/G12:C3 and G2:C13*/C3*:G12* sites in CPX1 and CPX1*, respectively (Figure 6B front view), at an angle of ca 62° between two PXZ moieties (Figure 6B side view). A close-up view of the crossover junction shows that the flipped A1 and G2 bases form complementary Watson–Crick base pairings with the symmetry-related T14* and C13* nucleotides at the ActD intercalation site. The strong 5'-GpC preference of ActD has preserved binding site interactions in the four-way-junction structure, with hydrogen bonding and stacking interactions between the PXZ ring and the THR of ActD and the guanine bases flanking the ring (Figure 6C and D). In this four-way junction structure, the DNA atoms comprising the formation of a crossover are separated by ca 3.8 Å, which further increases the stability of the superstructure. The base pair rise between G2 and the symmetry related C11* is ca 6.8 Å, which is significantly lower than the average rise distance of 7.7 Å caused by the PXZ intercalation site, suggesting that the formation of higher order structure may enhance the stacking between PXZ and DNA nucleotides to stabilize the overall junction structure.
Figure 6.
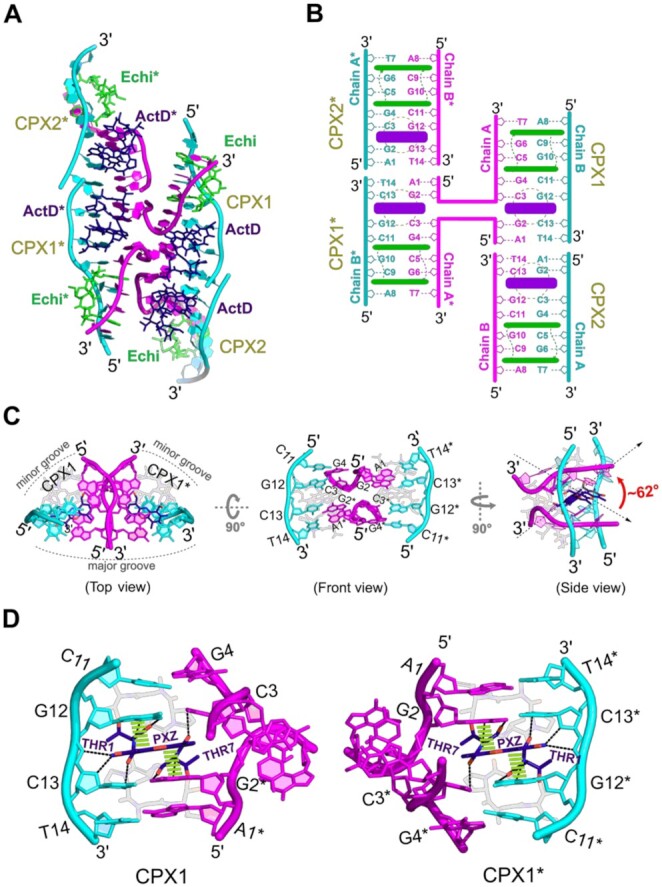
(A) Overall biological assembly of a discontinuous four-way junction superstructure formed by two independent duplexes of the G:C base pair complex structure through symmetrically related duplexes. The outer strands in the four-way junction structure are coloured in cyan, while the inner chains forming a crossover junction are shown in pink cartoons. The ActD and Echi are shown in blue and green sticks. Asterisk (*) symbols indicate symmetry-related complexes or drug molecules. (B) Schematic diagram of the superstructure of the four-way junction with detailed numbering of residues and insertion sites for ActD (blue) and Echi (green). (C) The core of the superstructure formed by the intersection of two symmetry-related complexes (CPX1 and CPX1*), viewed from above and from the front. A side view of the crossover junction shows that the two ActD molecules are arranged at an angle of about 62° to the plane of the phenoxazone (PXZ) rings. (D) Detailed hydrogen bonding (black dotted lines) and stacking interactions (green dashed lines) of PXZ and THR residues of ActD with G12pC13/G2*pC3 residues stabilize the junction site.
DISCUSSION
Specific DNA-targeting agents are one of the most challenging classes of compounds for cancer drug discovery: can one enhance cancer cell potency while reducing normal cell damage and resistance? Barton et al. have demonstrated the potential for targeted therapies in this field with metal intercalators to treat MMR-deficient tumours by targeting mismatch-containing DNA sequences (8,48). We have also previously reported several small molecule compounds (49–51) that have the potential to recognize various abnormal DNA sequences (such as mismatches and repeat-related structures). Understanding atomic details and the DNA interactions of small molecules may open up new opportunities for structure-based drug design to target specific disease-related DNA structures (52,53). A major caveat, however, is that the structural basis for the anticancer activity of DNA-targeting compounds has been primarily studied using one drug at a time. It has been postulated that a combination of two or more intercalators could bind tightly to distinct yet specific sites on DNA, thereby extending their sequence specificity (11). As far as we know, no study to date has provided a structural basis for targeting the same DNA structure simultaneously with two different anticancer drugs. As a start to elucidating the mechanism of such a DNA-targeted drug combination, we have determined the detailed ternary structures of two purine-related mismatched as well as two Watson–Crick base pair containing DNA sequences complexed with two well-known anticancer drugs, ActD and Echi. The distortion of the DNA backbone is affected by the action of the drug. Intercalation of two different drugs into one DNA sequence results in only one strand being significantly distorted, forming an orthogonal backbone shape. This suggests that DNA sequence is critical for the action of these drugs. Comparison of known ActD and Echi complexes bound separately to DNA duplexes with the current two-drug complexes shows that the general binding modes of ActD and Echi are conserved, but there are major differences in the changes to the DNA backbone (54–57). In contrast to the asymmetric backbone distortions reported here, the individual ActD and Echi complexes show symmetric backbone distortions of the two DNA strands. The simultaneous intercalation of ActD and Echi molecules can alleviate the steric conflict between the cyclic peptide units in the minor groove and absorbs torsional stress to compensate for the energetic cost required to unwind the backbone.
Out of the eight different types of DNA mismatches, the purine-related G:A mismatch is the most polymorphic, and can significantly destabilize duplex DNA. The occurrence of G:A mismatches has been detected in the pericentromeric regions of human chromosomes and in several single-stranded DNA viral genomes (58,59). In addition, these mismatches are also found in the noncanonical structures formed in (TGGAA)n repeats, which are known to cause the neurodegenerative disorder spinocerebellar ataxia 31 (SCA31) (44). Despite their biological importance, G:A mismatches have rarely been studied and structurally characterized using X-ray crystallography. The extent of structural disruption caused by mismatches or modified base pairs on DNA is highly dependent on sequence context (60,61). In the present study, we have confirmed that base-pair interchange in the case of an A:G to G:A mismatch leads to multiple mismatch conformations, including anti-anti, syn-anti, and a novel syn-like anti–anti geometry, which may cause differences in DNA stability. In the A:G mismatch complex, the central A4anti:G11anti mismatch forms two hydrogen-bond interactions. The sheared G4syn:A11anti and G4syn-like-anti:A11anti mismatches in the G:A complex results in a single hydrogen-bond interaction. Such sheared G:A mismatches are usually found in tandems in regular DNA duplexes (62,63). However, the current G:A complex shows that a single sheared G:A pairing can be observed when complexed with ActD and Echi. Moreover, the formation of Hoogsteen base pairs in DNA potentially plays an important role in DNA damage and repair (64–66). Several X-ray crystallography studies have reported the formation of Hoogsteen pairs upon small molecule binding, highlighting their importance as a specific recognition motif in DNA for small molecule ligands (67–69). We report here that the canonical C4anti:G11anti base pair at the two-drugs binding interface results in a Watson–Crick conformation after ActD and Echi binding. Interestingly, this canonical Watson–Crick pairing switches to a non-canonical Hoogsteen type when the central pair is changed to a Gsyn:Canti arrangement. The formation of such Hoogsteen base pairs at the two-drug binding interface also accounts for the reduction of the C1'-C1' distance by about 1.4 Å and increased intermolecular van der Waals interactions between the nucleotides and the drug molecules, which may result in additional stability to the complex structure compared to the Watson–Crick pair. These observations are consistent with several previously reported protein–DNA complexes, suggesting an analogous mechanism of action for these drug–DNA complexes (70).
The alteration of base pairing at the ligand binding interface may also have important implications for the recognition of DNA by drug or protein. For example, Al-Hashimi's group found that Echi or ActD can affect the rate of Hoogsteen base pair formation in Watson–Crick DNA duplexes at the drug binding interface (71). Indeed, our crystal structures containing a central C4:G11 pair, a G4:C11 pair, as well as G4:A11 and A4:G11 mismatches, also demonstrate that simultaneous intercalation of ActD and Echi can trap anti-anti-Watson–Crick pairing, syn-anti-Hoogsteen pairing or ‘syn-like’ anti–anti (sugar-edged) geometries in these complexes. A detailed comparison within these base pairs has revealed that when the QUI0 intercalation is oriented toward a non-guanine base, such as cytosine, or adenine (i.e. C4 or A4 base in the C:G and A:G complexes), a typical anti-anti conformation is well tolerated. When these structural features were compared, a strong correlation was observed between the geometry of the central base pairs and the DNA backbone deformation upon intercalation of two drugs (Table 1). In the A:G and C:G complexes, when the central base pair at the two drug-binding interface adopts a typical anti–anti or syn–anti geometry (in the case of G:A-CPX2) with a wobble or Watson–Crick conformation, the DNA forms a single-stranded ‘chair-like’ backbone deformation (chain B), and the complementary strand (chain A) remains straight and aligned to the DNA helix. Interestingly, in all these complex structures, the intercalation orientations of the ActD-PXZ and Echi-QUI0 moieties are parallel to each other (Supplementary Figure S10). Furthermore, the bis-intercalation of Echi results in a significant tilt between the planes of intercalated base pairs (Supplementary Figure S11). The N-methyl and βC-methyl side chains of MVA and ALA point toward C5 and G6 on one side and C9 and G10 on the other side, respectively, forming a wedge between the sugars of the polynucleotide. This introduces a significant tilt between the C5 (C9) and G6 (G10) planes and Echi-QUI0, respectively. This arrangement results in a decrease in the interplanar spacing between the unsaturated ring systems, leading to an increase in the successive stacking interactions between G2-PXZ-G12 at the ActD binding site and X4-QUI0-G10-C9 and A8-QUI1-G6-C5 residues at the Echi intercalation site. In long DNA molecules, such continuous triple stacking interactions can provide more intrinsic stability required for DNA, demonstrating the importance of single-stranded asymmetric backbone distortion when intercalating two drugs. In contrast to these observations, a more diverse DNA conformation occurs when the central base pair adopts a different geometry. For example, in G:A-CPX2, the central G4:A11 mismatch forms a new sugar-edged geometry, causing a sharp bend in the two DNA backbones to form a ‘curved’ duplex structure. The presence of a guanine (i.e. the G4 base) in the Echi-QUI0 ring in the G:C complex could cause a steric conflict between QUI0 and the bulky guanine base, forcing the guanine to adopt another conformation, i.e. the syn-conformation, forming a unique Hoogsteen base pairing. Previous crystal structures of Echi bound to oligomer DNA duplexes also exhibit Hoogsteen base pairs for the bases flanking the bis-intercalation site, suggesting that these play a significant role in inducing alternate base conformations (56,72). The formation of a central Hoogsteen base pairing also leads to more diverse DNA conformations, including kinked duplexes and a four-way junction superstructure. In these complexes, the ActD and Echi molecules show slanted intercalation towards the major groove, whereas the planar DNA bases and Echi-QUI0 are less inclined. These unusual arrangements resulted in reduced intermolecular stacking as well as other interactions such as hydrogen bonding. Thus, the differences in the intercalation angles and orientations of ActD and Echi seem to play an important role in binding to DNA base pairs, which accounts for the structural polymorphism caused by the two intercalators.
Table 1.
Structural feature comparison for all crystal structure complexes reported in the present study
| Homopurine mismatches | Watson–Crick base pairs | |||||
|---|---|---|---|---|---|---|
| Complex name | A:G | G:A-CPX1 | G:A-CPX2 | C:G | G:C-CPX1 | G:C-CPX2 |
| Central base-pair | A4:G11 | G4:A11 | G4:A11 | C4:G11 | G4:C11 | G4:C11 |
| Central base-pair geometry | anti–anti | syn–anti | ‘syn’ like anti–anti | anti–anti | syn–anti | syn–anti |
| Central base-pair type | Wobble | Wobble | Sugar-edged | Watson–Crick | Hoogsteen | Hoogsteen |
| Central base-pair C1'–C1' distance (Å) | 12.3 | 10.5 | 9.0 | 10.8 | 9.4 | 8.8 |
| ActD and Echi intercalation type | Parallel | Parallel | Slanted | Parallel | Parallel | Slanted |
| DNA conformation | Single strand distortion | Single strand distortion | Sharp bend | Single strand distortion | Four-way junction | Kinked |
| Backbone shape | ‘Chair’ shape | ‘Chair’ shape | ‘Curve’ shape | ‘Chair’ shape | Antiparallel crossover | ‘Zig-zag’ shaped |
| ActD | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Echi | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ |
| PDB ID | 7X6R | 7XDJ | 7X97 | 7X9F | ||
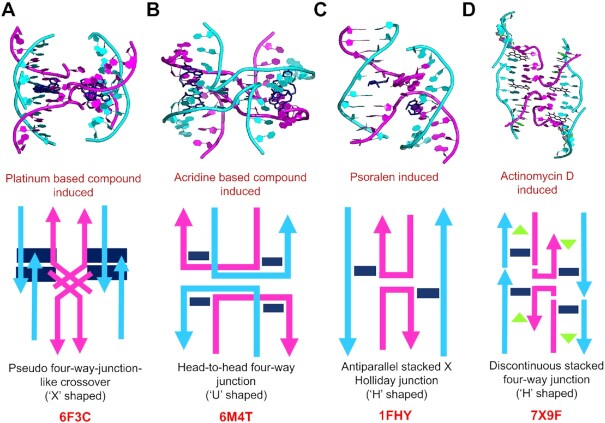
The induction of higher-order superstructures has been demonstrated in many nucleic acid-intercalator complexes, with potential applications in disease diagnosis and therapy (73–75). In a study by van Rixel et al. the ability of a small platinum compound [Pt(H2bapbpy)]-(PF6)2 (where H2bapbpy is N-(6-(6-(pyridin-2-ylamino)-pyridin-2-yl)-pyridin-2-yl)pyridin-2-amine) to induce a pseudo four-way junction like crossover through stacking and hydrogen bonding was identified, resulting in antiproliferative effects against various cancer cell lines (Figure 7A) (76). We have also reported a unique ‘U’ shaped head-to-head four-way junction formed by mismatch-containing DNA, which explains the supramolecular chemistry of triaminotriazine DNA intercalators, inducing higher-order superstructure with applications in drug design (Figure 7B) (77). Further, Eichman et al. showed that a psoralen based DNA intercalator drug can form a typical stacked antiparallel Holliday junction by cross-linking with thymine bases, which can be relevant to the repair of psoralen-lesions in mammalian DNA (Figure 7C) (78). In the current study, we found that a complex comprising a central G:C Hoogsteen base pair could induce a discontinuous and unique antiparallel four-way junction-like higher-order structure at the ActD intercalation site (Figure 7D). After comparing these four-way junction structures, we found that two ActD molecules bound across the centre of the junction to form a peculiar topology, in some ways similar to a typical Holiday junction structure, in which the flipped-out bases form canonical Watson–Crick base pairing flanking the crossover. ActD has been previously found to induce many different structural changes in DNA, such as base-flipped backbone kinks and the formation of pseudo intercalated complexes (79–82). Our crystal structure has shown, for the first time, that ActD is equally capable of introducing a four-way junction-like superstructure. The crossover at the 5'-terminal end begins near G4 in ‘chain A’, which adopts a syn conformation, suggesting a potential link between the Hoogsteen base pair and the formation of a four-way junction-like structures. It would be interesting to investigate the role of Hoogsteen base pairs in the induction of such higher-order structures mediated by small molecule ligands, which may have potential biological and biotechnological applications.
Figure 7.
Comparison of DNA topologies of four-way junction superstructures in different sequences induced by intercalators. (A) The ‘X’-shaped crossover topology generated in a pseudo four-way junction superstructure induced by a platinum-based [Pt(H2bapbpy)]-(PF6)2 compound. (B) When a triaminotriazine DNA intercalator is inserted into the T:T mismatch duplex, a ‘U’-shaped head-to-head four-way junction like topology is formed. (C) Psoralen-based 4'-hydroxymethyl-4,5',8-trimethylpsoralen (HMT) DNA intercalation compound induces a Holliday junction by cross-linking with thymine bases to form an antiparallel stacked topology. (D) Intercalation of ActD induces a discontinuous four-way junction with an antiparallel alignment of the DNA backbone strands. Cyclic peptide parts of ActD and Echi has been removed to enhanced clarity.
Our results underscore the importance of base pair modulation by small molecules during DNA recognition for the development of new anticancer strategies. Such novel DNA changes, distinct from the structurally simpler changes produced by classic intercalating drugs such as ActD or doxorubicin, could well reduce resistance, as well as enhancing therapeutic indices based on selective drug interaction at DNA lesions induced by cancer-related mutagenesis. The polymorphic structures presented in this study are clearly a consequence of base pair interchange effects and are not the result of crystal packing forces and are thus likely to represent their biologically relevant conformations when two drugs bind to DNA. The combination of two or more DNA intercalators may also be an effective approach for the treatment for DNA mismatch-related cancers. Based on our structural observations, we have established a working hypothesis for using the combination of ActD and Echi against MMR-deficient cancers, even though a detailed linkage between our structural observations and the biological data remains to be fully elucidated. The fact that the two simultaneous drug binding results in unexpected and unprecedented structural changes to the target DNA sequence is consistent with the concept that the resulting DNA lesions are more vulnerable in MMR-deficient cells and tumours. We have now shown experimentally that the combination of the two drugs can synergistically enhance cytotoxicity (Satange et al., 2022, under review) in MMR-deficient HCT116 cells. We have also shown that synergistic in vivo therapy with ActD and Echi can minimize off-target and generalized cytotoxicity and maximize anti-cancer efficacy even at 10-fold lower doses of a single drug in a xenograft mouse model for MMR-deficient tumours. We therefore suggest that the combination of two DNA intercalators may have clinical potential as an effective and reduced toxicity chemotherapeutic strategy.
In summary, our results provide evidence of new structural features for two different DNA intercalators binding to DNA containing purine-purine G:A and A:G, and Watson–Crick G:C and C:G base pairs. We have identified a binding mechanism by which two different drugs induced specific base-pairing changes in DNA. Determining various drug-DNA complex structures of this type could help in understanding the nature of various specific and non-specific intermolecular interactions that contribute to the stabilization of mismatches in DNA through specific backbone alterations. The structural features discussed here may be also generalized for other drug–DNA and protein–DNA complex structures. It is hoped that this study will help guide the development of mismatch binding agents for future generations of DNA-targeted chemotherapy.
DATA AVAILABILITY
The atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank with accession codes 7X6R, 7XDJ, 7X9F and 7X97.
Supplementary Material
ACKNOWLEDGEMENTS
Authors sincerely thank the National Synchrotron Radiation Research Center (Taiwan) staff for their help in X-ray data collection.
Contributor Information
Roshan Satange, Institute of Genomics and Bioinformatics, National Chung Hsing University, Taichung 402, Taiwan; Ph.D. Program in Medical Biotechnology, National Chung Hsing University, Taichung 402, Taiwan.
Shih-Hao Kao, Institute of Biotechnology, National Chung Hsing University, Taichung 402, Taiwan.
Ching-Ming Chien, Institute of Genomics and Bioinformatics, National Chung Hsing University, Taichung 402, Taiwan.
Shan-Ho Chou, Institute of Biochemistry, National Chung Hsing University, Taichung 402, Taiwan.
Chi-Chien Lin, Institute of Biomedical Science, National Chung Hsing University, Taichung 402, Taiwan.
Stephen Neidle, The School of Pharmacy, University College London, London WC1N 1AX, United Kingdom.
Ming-Hon Hou, Institute of Genomics and Bioinformatics, National Chung Hsing University, Taichung 402, Taiwan; Ph.D. Program in Medical Biotechnology, National Chung Hsing University, Taichung 402, Taiwan; Institute of Biotechnology, National Chung Hsing University, Taichung 402, Taiwan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan, R.O.C. [109-2628-M-005-001-MY4, 109-2311-B-005-007-MY3 to M.-H.H.]; joint grant of National Chung Hsing University and Chung Shan Medical University [NCHU-CSMU 10904, NCHU-CSMU 10803 to M.-H.H.]. Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Granzhan A., Kotera N., Teulade-Fichou M.-P.. Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014; 43:3630–3665. [DOI] [PubMed] [Google Scholar]
- 2. Gibson D. Drug–DNA interactions and novel drug design. Pharmacogenomics J. 2002; 2:275–276. [DOI] [PubMed] [Google Scholar]
- 3. Satange R., Chang C.-K., Hou M.-H.. A survey of recent unusual high-resolution DNA structures provoked by mismatches, repeats and ligand binding. Nucleic Acids Res. 2018; 46:6416–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Portugal J., Barceló F.. Noncovalent binding to DNA: still a target in developing anticancer agents. Curr. Med. Chem. 2016; 23:4108–4134. [DOI] [PubMed] [Google Scholar]
- 5. Rescifina A., Zagni C., Varrica M.G., Pistarà V., Corsaro A.. Recent advances in small organic molecules as DNA intercalating agents: synthesis, activity, and modeling. Eur. J. Med. Chem. 2014; 74:95–115. [DOI] [PubMed] [Google Scholar]
- 6. Paul A., Bhattacharya S.. Chemistry and biology of DNA-binding small molecules. Curr. Sci. 2012; 102:212–231. [Google Scholar]
- 7. Martins-Teixeira M.B., Carvalho I.. Antitumour anthracyclines: progress and perspectives. ChemMedChem. 2020; 15:933–948. [DOI] [PubMed] [Google Scholar]
- 8. Boyle K.M., Barton J.K.. Targeting DNA mismatches with rhodium metalloinsertors. Inorg. Chim. Acta. 2016; 452:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tandon R., Luxami V., Kaur H., Tandon N., Paul K.. 1,8-Naphthalimide: A potent DNA intercalator and target for cancer therapy. Chem. Rec. 2017; 17:956–993. [DOI] [PubMed] [Google Scholar]
- 10. Brana M.F., Cacho M., Gradillas A., Pascual-Teresa B., Ramos A.. Intercalators as anticancer drugs. Curr. Pharm. Des. 2001; 7:1745–1780. [DOI] [PubMed] [Google Scholar]
- 11. Takusagawa H.L., Takusagawa F.. Crystallization and preliminary X-ray diffraction studies of d(ACGTAGCTACGT)2:[actinomycin D, (echinomycin)2] and d(ACGTAGCTACGT)2:[actinomycin D, (triostin A)2] complexes. Acta Crystallogr. D Biol. Crystallogr. 2000; 56:344–347. [DOI] [PubMed] [Google Scholar]
- 12. Weidmann A.G., Barton J.K.. A monofunctional platinum complex coordinated to a rhodium metalloinsertor selectively binds mismatched DNA in the minor groove. Inorg. Chem. 2015; 54:9626–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orson F.M., Kinsey B.M., McShan W.M.. Linkage structures strongly influence the binding cooperativity of DNA intercalators conjugated to triplex forming oligonucleotides. Nucleic Acids Res. 1994; 22:479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma L., Ma R., Wang Z., Yiu S.-M., Zhu G.. Heterodinuclear Pt(iv)–Ru(ii) anticancer prodrugs to combat both drug resistance and tumor metastasis. Chem. Commun. 2016; 52:10735–10738. [DOI] [PubMed] [Google Scholar]
- 15. Waring M.J., Wakelin L.P.G.. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974; 252:653–657. [DOI] [PubMed] [Google Scholar]
- 16. Ughetto G., Wang A.H.J., Quigley G.J., van der Marel G.A., van Boom J.H., Rich A.. A comparison of the structure of echinomycin and triostin a complexed to a DNA fragment. Nucleic Acids Res. 1985; 13:2305–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamitori S., Takusagawa F.. Multiple binding modes of anticancer drug actinomycin D: X-ray, molecular modeling, and spectroscopic studies of d(GAAGCTTC)2-Actinomycin D complexes and its host DNA. J. Am. Chem. Soc. 1994; 116:4154–4165. [Google Scholar]
- 18. Koba M., Konopa J.. [Actinomycin d and its mechanisms of action]. Postepy Hig. Med. Dosw. (Online). 2005; 59:290–298. [PubMed] [Google Scholar]
- 19. Yun J., Lee S.W., Lim S.H., Kim S.H., Kim C.K., Park S.K.. Successful treatment of a high-risk nonseminomatous germ cell tumor using etoposide, methotrexate, actinomycin D, cyclophosphamide, and vincristine: a case report. World J. Clin. Cases. 2020; 8:5334–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas A., Samykutty A., Gomez-Gutierrez J.G., Yin W., Egger M.E., McNally M., Chuong P., MacCuaig W.M., Albeituni S., Zeiderman M.et al.. Actively targeted nanodelivery of echinomycin induces autophagy-mediated death in chemoresistant pancreatic cancer in vivo. Cancers (Basel). 2020; 12:2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schilsky R.L., Faraggi D., Korzun A., Vogelzang N., Ellerton J., Wood W., Henderson I.C.. Phase II study of echinomycin in patients with advanced breast cancer: a report of cancer and leukemia group b protocol 8641. Invest. New Drugs. 1991; 9:269–272. [DOI] [PubMed] [Google Scholar]
- 22. Albertini V., Jain A., Vignati S., Napoli S., Rinaldi A., Kwee I., Nur-e-Alam M., Bergant J., Bertoni F., Carbone G.M.et al.. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer streptomyces argillaceus exhibits improved transcriptional repressor activity: implications for cancer therapy. Nucleic Acids Res. 2006; 34:1721–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan J., Zhu L., Wang B.. From GC-rich DNA binding to the repression of survivin gene for quercetin nickel (II) complex: implications for cancer therapy. Biometals. 2010; 23:1075–1084. [DOI] [PubMed] [Google Scholar]
- 24. Beniaminov A.D., Chashchina G.V., Livshits M.A., Kechko O.I., Mitkevich V.A., Mamaeva O.K., Tevyashova A.N., Shtil A.A., Shchyolkina A.K., Kaluzhny D.N.. Discrimination between G/C binding sites by olivomycin a is determined by kinetics of the Drug-DNA interaction. Int. J. Mol. Sci. 2020; 21:5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fryxell K.J., Moon W.-J.. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 2004; 22:650–658. [DOI] [PubMed] [Google Scholar]
- 26. Behringer M.G., Hall D.W.. Genome-wide estimates of mutation rates and spectrum in schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda). 2015; 6:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan F., Man V.H., Roland C., Sagui C.. Structure and dynamics of DNA and RNA double helices obtained from the CCG and GGC trinucleotide repeats. J. Phys. Chem. B. 2018; 122:4491–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rossetti G., Dans P.D., Gomez-Pinto I., Ivani I., Gonzalez C., Orozco M.. The structural impact of DNA mismatches. Nucleic Acids Res. 2015; 43:4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan F., Zhang Y., Man V.H., Roland C., Sagui C.. E-motif formed by extrahelical cytosine bases in DNA homoduplexes of trinucleotide and hexanucleotide repeats. Nucleic Acids Res. 2017; 46:942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Roland C., Sagui C.. Structure and dynamics of DNA and RNA double helices obtained from the GGGGCC and CCCCGG hexanucleotide repeats that are the hallmark of C9FTD/ALS diseases. ACS Chem. Neurosci. 2017; 8:578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maity A., Winnerdy F.R., Chen G., Phan A.T.. Duplexes formed by G4C2 repeats contain alternate slow- and fast-flipping G·G base pairs. Biochemistry. 2021; 60:1097–1107. [DOI] [PubMed] [Google Scholar]
- 32. Sathyamoorthy B., Shi H., Zhou H., Xue Y., Rangadurai A., Merriman D.K., Al-Hashimi H.M.. Insights into Watson–Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res. 2017; 45:5586–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mukherjee S., Błaszczyk L., Rypniewski W., Falschlunger C., Micura R., Murata A., Dohno C., Nakatani K., Kiliszek A.. Structural insights into synthetic ligands targeting A–A pairs in disease-related CAG RNA repeats. Nucleic Acids Res. 2019; 47:10906–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ni L., Yamada T., Nakatani K.. Assembly of ruthenium complexes on double stranded DNA using mismatch binding ligands. Chem. Commun. 2020; 56:5227–5230. [DOI] [PubMed] [Google Scholar]
- 35. Li Y., Zon G., Wilson W.D.. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997; 276:Academic Press; 307–326. [DOI] [PubMed] [Google Scholar]
- 37. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laskowski R.A., Swindells M.B.. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011; 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- 40. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li S., Olson W.K., Lu X.-J.. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019; 47:W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satange R., Chuang C.-Y., Neidle S., Hou M.-H.. Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation. Nucleic Acids Res. 2019; 47:8899–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allawi H.T., SantaLucia J.. Nearest neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry. 1998; 37:2170–2179. [DOI] [PubMed] [Google Scholar]
- 44. Huang T.-Y., Chang C.-k., Kao Y.-F., Chin C.-H., Ni C.-W., Hsu H.-Y., Hu N.-J., Hsieh L.-C., Chou S.-H., Lee I.-R.et al.. Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliveira L.M., Long A.S., Brown T., Fox K.R., Weber G.. Melting temperature measurement and mesoscopic evaluation of single, double and triple DNA mismatches. Chem. Sci. 2020; 11:8273–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kan L.S., Chandrasegaran S., Pulford S.M., Miller P.S.. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:4263–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chou S.-H., Zhu L., Reid B.R.. Sheared purine·purine pairing in biology. J. Mol. Biol. 1997; 267:1055–1067. [DOI] [PubMed] [Google Scholar]
- 48. Threatt S.D., Synold T.W., Wu J., Barton J.K.. In vivo anticancer activity of a rhodium metalloinsertor in the HCT116 xenograft tumor model. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:17535–17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tseng W.-H., Chang C.-k., Wu P.-C., Hu N.-J., Lee G.-H., Tzeng C.-C., Neidle S., Hou M.-H.. Induced-Fit recognition of CCG trinucleotide repeats by a nickel–chromomycin complex resulting in large-scale DNA deformation. Angew. Chem. Int. Ed. 2017; 56:8761–8765. [DOI] [PubMed] [Google Scholar]
- 50. Lo Y.-S., Tseng W.-H., Chuang C.-Y., Hou M.-H.. The structural basis of actinomycin D–binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013; 41:6782–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen F.-M., Sha F., Chin K.-H., Chou S.-H.. The nature of actinomycin d binding to d(AACCAXYG) sequence motifs. Nucleic Acids Res. 2004; 32:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Addess K.J., Feigon J.. NMR investigation of Hoogsteen base pairing in quinoxaline antibiotic–DNA complexes: comparison of 2:1 echinomycin, triostin A and [N-MeCys3,N-MeCys7] TANDEM complexes with DNA oligonucleotides. Nucleic Acids Res. 1994; 22:5484–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chin K.H., Chen F.M., Chou S.H.. Solution structure of the Actd–5′-CCGTT3GTGG-3′ complex: drug interaction with tandem G·T mismatches and hairpin loop backbone. Nucleic Acids Res. 2003; 31:2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu P.-C., Tzeng S.-L., Chang C.-k., Kao Y.-F., Waring M.J., Hou M.-H.. Cooperative recognition of T:T mismatch by echinomycin causes structural distortions in DNA duplex. Nucleic Acids Res. 2018; 46:7396–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hou M.H., Robinson H., Gao Y.G., Wang A.H.J.. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002; 30:4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cuesta-Seijo J.A., Sheldrick G.M.. Structures of complexes between echinomycin and duplex DNA. Acta Crystallogr. D Biol. Crystall. 2005; 61:442–448. [DOI] [PubMed] [Google Scholar]
- 57. Chen H., Liu X., Patel D.J.. DNA bending and unwinding associated with actinomycin D antibiotics bound to partially overlapping sites on DNA. J. Mol. Biol. 1996; 258:457–479. [DOI] [PubMed] [Google Scholar]
- 58. Jaishree T.N., Wang A.H.J.. Human chromosomal centromere (AATGG)n sequence forms stable structures with unusual base pairs. FEBS Lett. 1994; 347:99–103. [DOI] [PubMed] [Google Scholar]
- 59. Shlomai J., Kornberg A.. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Voigt J.M., Topal M.D.. O6-Methylguanine and A:C and G:T mismatches cause asymmetric structural defects in DNA that are affected by DNA sequence. Biochemistry. 1990; 29:5012–5018. [DOI] [PubMed] [Google Scholar]
- 61. Mondal M., Yang L., Cai Z., Patra P., Gao Y.Q.. A perspective on the molecular simulation of DNA from structural and functional aspects. Chem. Sci. 2021; 12:5390–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gao Y.-G., Robinson H., Sanishvili R., Joachimiak A., Wang A.H.J.. Structure and recognition of sheared tandem G·A base pairs associated with human centromere DNA sequence at atomic resolution. Biochemistry. 1999; 38:16452–16460. [DOI] [PubMed] [Google Scholar]
- 63. Neidle S., Sanderson M.. Neidle S., Sanderson M.. Chapter 4: Non-standard and higher-order DNA structures: DNA-DNArecognition. Principles of Nucleic Acid Structure. 2022; 2nd ednNY: Academic Press; 109–190. [Google Scholar]
- 64. Kitayner M., Rozenberg H., Rohs R., Suad O., Rabinovich D., Honig B., Shakked Z.. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2010; 17:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aishima J., Gitti R.K., Noah J.E., Gan H.H., Schlick T., Wolberger C.. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002; 30:5244–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shi H., Kimsey I.J., Gu S., Liu H.-F., Pham U., Schumacher MA., Al-Hashimi HM.. Revealing A-T and G-C Hoogsteen base pairs in stressed protein-bound duplex DNA. Nucleic Acids Res. 2021; 49:12540–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Quigley G.J., Ughetto G., Marel G.A.v.d., Boom J.H.v., Wang A.H.-J., Rich A.. Non-Watson–Crick G.C and A.T base pairs in a DNA-antibiotic complex. Science. 1986; 232:1255–1258. [DOI] [PubMed] [Google Scholar]
- 68. Singh U.C., Pattabiraman N., Langridge R., Kollman P.A.. Molecular mechanical studies of d(CGTACG)2: complex of triostin A with the middle A-T base pairs in either Hoogsteen or Watson-Crick pairing. Proc. Natl. Acad. Sci. 1986; 83:6402–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gao X., Patel D.J.. NMR studies of echinomycin bisintercalation complexes with d(A1-C2-G3-T4) and d(T1-C2-G3-A4) duplexes in aqueous solution: sequence-dependent formation of Hoogsteen A1.T4 and Watson-Crick T1.A4 base pairs flanking the bisintercalation site. Biochemistry. 1988; 27:1744–1751. [DOI] [PubMed] [Google Scholar]
- 70. Afek A., Shi H., Rangadurai A., Sahay H., Senitzki A., Xhani S., Fang M., Salinas R., Mielko Z., Pufall M.A.et al.. DNA mismatches reveal conformational penalties in protein–DNA recognition. Nature. 2020; 587:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu Y., McSally J., Andricioaei I., Al-Hashimi H.M.. Modulation of Hoogsteen dynamics on DNA recognition. Nat. Commun. 2018; 9:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cuesta-Seijo J.A., Weiss M.S., Sheldrick G.M.. Serendipitous SAD phasing of an echinomycin-(ACGTACGT)2 bisintercalation complex. Acta Crystallog. D Biol. Crystallogr. 2006; 62:417–424. [DOI] [PubMed] [Google Scholar]
- 73. Zell J., Duskova K., Chouh L., Bossaert M., Chéron N., Granzhan A., Britton S., Monchaud D.. Dual targeting of higher-order DNA structures by azacryptands induces DNA junction-mediated DNA damage in cancer cells. Nucleic Acids Res. 2021; 49:10275–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Howell L.A., Waller Z.A.E., Bowater R., O’Connell M., Searcey M.. A small molecule that induces assembly of a four way DNA junction at low temperature. Chem. Commun. 2011; 47:8262–8264. [DOI] [PubMed] [Google Scholar]
- 75. Kolb F.A., Malmgren C., Westhof E., Ehresmann C., Ehresmann B., Wagner E.G.H., Romby P.. An unusual structure formed by antisense-target RNA binding involves an extended kissing complex with a four-way junction and a side-by-side helical alignment. RNA. 2000; 6:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Rixel V.H.S., Busemann A., Wissingh M.F., Hopkins S.L., Siewert B., van de Griend C., Siegler M.A., Marzo T., Papi F., Ferraroni M.et al.. Induction of a four-way junction structure in the DNA palindromic hexanucleotide 5′-d(CGTACG)-3′ by a mononuclear platinum complex. Angew. Chem. Int. Ed. 2019; 58:9378–9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chien C.-M., Wu P.-C., Satange R., Chang C.-C., Lai Z.-L., Hagler L.D., Zimmerman S.C., Hou M.-H.. Structural basis for targeting T:T mismatch with triaminotriazine-acridine conjugate induces a U-shaped head-to-head four-way junction in CTG repeat DNA. J. Am. Chem. Soc. 2020; 142:11165–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eichman B.F., Mooers B.H.M., Alberti M., Hearst J.E., Ho P.S.. The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions. J. Mol. Biol. 2001; 308:15–26. [DOI] [PubMed] [Google Scholar]
- 79. Lo Y.-S., Tseng W.-H., Chuang C.-Y., Hou M.-H.. The structural basis of actinomycin D–binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013; 41:4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chou S.-H., Chin K.-H., Chen F.-M.. Looped out and perpendicular: deformation of Watson–Crick base pair associated with actinomycin D binding. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Robinson H., Gao Y.-G., Yang X.-l., Sanishvili R., Joachimiak A., Wang A.H.J.. Crystallographic analysis of a novel complex of actinomycin D bound to the DNA decamer CGATCGATCG. Biochemistry. 2001; 40:5587–5592. [DOI] [PubMed] [Google Scholar]
- 82. Takusagawa F., Dabrow M., Neidle S., Berman H.M.. The structure of a pseudo intercalated complex between actinomycin and the DNA binding sequence d(GpC). Nature. 1982; 296:466–469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank with accession codes 7X6R, 7XDJ, 7X9F and 7X97.