Abstract
Mass-spectrometry-based proteomics presents some unique challenges for batch effect correction. Batch effects are technical sources of variation, can confound analysis and usually non-biological in nature. As proteomic analysis involves several stages of data transformation from spectra to protein, the decision on when and what to apply batch correction on is often unclear. Here, we explore several relevant issues pertinent to batch effect correct considerations. The first involves applications of batch effect correction requiring prior knowledge on batch factors and exploring data to uncover new/unknown batch factors. The second considers recent literature that suggests there is no single best batch effect correction algorithm---i.e., instead of a best approach, one may instead ask, what is a suitable approach. The third section considers issues of batch effect detection. And finally, we look at potential developments for proteomic-specific batch effect correction methods and how to do better functional evaluations on batch corrected data.
Keywords: Proteomics, Batch effects, Batch correction, Batch visualization
1. Introduction
Batch effects (BEs) are unwanted variation in data produced from technical sources such as the machine type and/or experimenter. BEs are sometimes referred to as technical bias. If undealt with or mishandled (using inappropriate correction methods), BEs may confound analysis, and lead towards mis-estimation of effect sizes (e.g., the magnitude of difference of protein expression level between different phenotypes). In more severe cases, it can lead towards false positives (proteins that are not differential are selected) and false negatives (proteins that are differential are not selected) [1]. In general, BEs are complex, and effective mitigation is highly context dependent [2], [3]. It is also surprising that even as technologies and analytical methods advance, BEs seem to become more pertinent and relevant [4].
Advancement of biomedical science is dependent on high throughput profiling of biological states. Being able to characterize the unique complement of genes, proteins and metabolites being expressed in samples of interest may allow us to understand the causal factors and functional characteristics underpinning a disease or phenotype. Since proteins are the functional units of cells, being able to assay the protein complement is especially critical. But unlike gene expression profiling (which is matured), proteome profiling (i.e., the complete set of proteins relevant to a phenotype) is ostensibly more challenging.
The current prevailing technology for proteome profiling is mass-spectrometry (MS)-based proteomics [5]. This is a powerful technology that involves a series of complex sample processing, data acquisition, and data analysis steps: Proteins are first extracted from samples and digested into smaller manageable fragments known as peptides. These peptides are then labelled and/or ionized (depending on the proteomic setup) and detected in the first MS dimension. This is known as MS1. Selected (or all isolatable) peptides are then fragmented into even smaller fragments, before being captured in a second MS dimension. This is known as MS2. Both MS1 and MS2 data are then combined and integrated to identify the peptides in a process known as peptide-spectrum matching. The identified peptides are then reassembled using protein assembly algorithms to produce proteins [6]. The afore-described procedures are typically known as tandem-MS, or MS/MS. Many other variations of this setup exist but in general, are no less complex nor involve fewer steps.
Mass-spectrometry-based proteomics presents some unique challenges for BE correction. The complexity of steps and processes in MS-based proteomics can introduce various levels and intermingling of technical biases and errors, therefore making effective BE correction difficult. For example, if peptides from different samples are analyzed on different MS machines, or if the digestion procedure involves different reagent lots [7], different levels of BEs are introduced into the data. Since each step may introduce different BEs, this means it is important to collect as much meta-information as possible. Information such as who handled which samples, and which machines were used to acquire spectra, are important, as such information are essentially batch factors that could be evaluated for non-negligible BEs. Conversely, such multi-tiered issues pertaining to BEs also means that there remains ample opportunity for creative and interesting BE correction strategies in proteomics. We discuss some interesting issues and potential solutions.
2. Do (Should) I know everything about my batch effect?
Many popular batch-effect correction algorithms (BECAs) e.g., ComBat [8] and Harman [9] require explicit input of batch factors i.e., how the data is divisible by batch. If prior knowledge on batch factors is not available, an alternative is to use Surrogate Variable Analysis (SVA) [10], [11]. SVA estimate information correlated with class information (i.e., the biological information we want to retain), treats all non-correlated sources of variation as batch, and removes them accordingly. One may imagine such an approach while powerful, is not very satisfying. Suppose if class information is modulated and/or correlated with other sources of variation, SVA may overcorrect. Jaffe et al. reported that by defining the biological effect of interest to be the average change in expression with treatment, SVA removes many individual sample-specific expression traits and even secondary effects of interest [12].
Hence, to use SVA well, one needs to know how to precisely define and carefully specify biological effects of interest. If we are interested in exploring the impact of additional factors on the outcome of interest, then these additional factors would also have to be specified in the SVA model [12]. Unfortunately, this is not a straightforward process. Also, to explore these additional factors, prior knowledge on the structure of these additional factors is required. If these additional factors are not specified (or known), then they must be inferred.
And what can be done if batch effects are suspected but not known? The BatchI approach offers an interesting possibility [13]. BatchI attempts to dynamically discover batch structures or batch factors by partitioning a series of data (e.g., proteomics expression matrices) into sub-series corresponding to estimated batches. Estimated batches in turn, are based on attempts to split data with maximal dispersion between batches within maintaining minimal within batch dispersion. This approach allows us to explore and understand our data better by discovering unknown/unreported BEs. This information is very helpful as it may pin-point potential flaws in our experimental processes. Whilst BEs should be removed eventually to help us advance our understanding of biology, understanding the sources and structure of BEs are also very important. BatchI is available online as an R package at https://kiiaed.aei.polsl.pl/index.php/pl/oprogramowanie-zaed.
3. Is there a best batch effect correction algorithm for proteomics data?
To date, many benchmark studies have been performed on high-throughput datasets across a variety of BECAs. These are mostly limited to bulk sequencing [14] and single-cell sequencing data [15] but some work has also been performed on proteomics datasets [3].
The results are varied: For example, Luo et al’s study on microarray gene expression data suggests that ratio-based are superior especially on imbalanced data [14]. We conducted a similar battery of BECAs evaluated on proteomics data but found otherwise [3]. In our evaluation, only ComBat was able to perform adequately well for imbalanced data, while SVA and Harman suffered heavily. However, we also note the performance of BECAs are dependent on data innate characteristics, but also compatibilities with normalization and data transformation approaches [3].
There may not be a universal best batch correction approach. Instead, we advocate it is first important to study and explore the data itself first. This can be done via simple exploratory approaches such as the side-by-side boxplots for visualizing correlations of batch effects with principal components on data [16], and also the side-by-side barcharts for investigating gene-gene correlations [7]. After which, we may then propose the most appropriate BECA for the data. To achieve this, it is important to understand the assumptions made by the BECA (e.g., does it treat BEs as constant or noisy? Additive, multiplicative or mixed?), and whether any prior data processing or normalization approach is compatible with these assumptions.
In proteomics data, there is an added consideration known as drift effects, which are particular to the nature of MS instrumentation. This is critical when dealing with experiments involving large sample sizes, typically in the order of hundreds [17]. Unlike traditional BEs which are dependent on specific (discrete) machines, reagent lots or experimenters, drift effects manifest as “continuous effects” over time (one way to imagine this is that time is a factor with many levels). When visible across hundreds of samples ordered by running sequence, drift effects can be adjusted by baseline correction or regression methods. One needs to be careful when performing this form of correction should there be multiple classes in data, especially if the data comes with very strong class effects. In such cases, it may be worthwhile distributing different class samples evenly across the running sequence to avoid biasing the correction.
If there is no perfect BECA and we know BECAs are also affected by other processing steps, then are there acceptable procedures or protocols for general use?
If there is no perfect BECA and we know BECAs are also affected by other processing steps, then are there acceptable procedures or protocols for general use? Interestingly in proteomics, we are only aware of one recently published set of principles or best practices by Cuklina et al [17] which integrated perspectives and insights from several other published works. Batch correction is ultimately not a straightforward procedure and should never be seen as such. Earlier, we stated that we should identify a BECA suitable for the data. That is one aspect, but another important point is to also align the batch correction process with the larger research goal. For example, if the goal is to correct data for subsequent use in a machine learning task, it would make sense to preserve much of the original data scale and data integrity (you would not want to change it greatly using methods such as SVA). It would also make sense not to use ratio-based batch correction methods as these would effectively merge information across classes, which makes classification tasks such as class prediction impossible.
If functional analysis is the goal, such that we are primarily concerned with advancing our knowledge of the underlying biology (following comparative analyses between samples of different classes (e.g., cancer versus normal)), then some useful approaches do exist, which do not necessary require batch correction. One approach is to use strong discretization normalization approaches---one example, the gene fuzzy scoring (GFS) method reduces each sample such that only proteins ranked in the top 10 % (based on abundance) are given a value of 1, those between 10 and 15 % are interpolated between 0 and 1, and those falling over 15 % are assigned a value of 0, and thus ignored [18]. The idea is that each sample can be uniquely represented by its top proteins, and that these top proteins should be fairly conserved amongst samples within the same class, but not in the opposing class. Since values are set between 0 and 1, BEs which alters expression counts will have lessened effects. We can extend the idea by representing each sample in terms of those networks and systems enriched for those top proteins with non-zero values [16]. Indeed, we found that network-based approaches may have some resistance against BEs. However, methods like GFS or networks are not panaceas and do come with cost. GFS is a brutal procedure resulting in massive loss of information: absolute expression information is converted into binaries, 80–85 % of proteins are converted to 0s if they are not top ranked in the tissue. But GFS is very stable and can produce similar results on feature selection even if we relax its cutoff parameters [19]. It is suitable for noisy or challenging data. Network-based approaches can further improve the reproducibility of methods such as GFS [16], but are constrained by the availability of high-quality network information.
In practice, many uses principal components analysis (PCA) scatterplots to visually inspect for the presence of BEs but go no further than that. This method can be extended systematically for the calculation of correlations between principal components (PCs) and BEs [19], [20]. PCs are projections of high-dimensional data that are orthogonal and thus, independent of each other. It serves as a very powerful dimensionality reduction method. But also can be used for dealing with BEs in a very simple and intuitive way: Suppose if the first PC (PC1) is the batch-correlated PC, we can simply drop it from analysis, and use the remaining PCs for functional and clustering analyses [18], [19], [20], [21], [22] (i.e., we treat the PCs like independent features directly).
4. When is the best time to remove my batch effects?
In Cuklina et al’s paper, there was considerable discussion on when is the best time to tackle BEs [17]. Proteomics comprises several steps, from spectra acquisition to peptide-spectra matching and finally protein assembly and quantitation. One can decide the most appropriate or strategic window to perform BE correction.
Unfortunately, there is little in current literature on optimal correction window for proteomics. But there are some interesting observations. Graw et al reported that in their studies, BE is more prevalent in the raw peptide data than in the filtered protein data [23]. This seems to suggest in part, some important batch information is lost during the transition from spectra to protein. Another related observation comes from Brenes et al, where they reported missing value inflation when attempting to integrate data from multiple batches [24]. They highlighted the issue is aggravated at the peptide level, which comes as no surprise. However, this may mean that seemingly lower missingness at the protein level is probably due to different peptides mappable to the same protein, found across different samples. If BE information is not consistently distributed across peptides for each protein, then this may lead also to subsequent errors in estimation and correction.
These observations do not guarantee early batch correction (e.g., at spectra or peptide level) brings better outcomes. Dealing with raw data also presents challenges: E.g., if we are dealing directly with spectra or peptides, we will also be dealing with higher dimensionality, noise and missingness issues as well.
So, do we need early correction? For most intents and purposes, late BE correction (at protein level appears) works reasonably well and is commonly practiced anyway. Does this therefore mean that we can safely do away with early BE correction?
We do not think so. Recently, we demonstrated that by deliberately mis-imputing missing data by borrowing information from other batches, we effectively convert structured batch variation into noise, increasingly the overall variability at the sample level, and increasing the chance of incurring false positives/negatives during functional analysis [25]. Although we did not perform these simulations at the peptide level, it is not unfathomable to think that mistakes incurred due to MVI at peptide level, would also have similar effects. Thus, we believe there is benefit in developing approaches that can help enhance peptide level imputation and support early BE correction. However, more development is needed.
5. Is there a reliable measure for batch effect besides looking at scatterplots?
PCA coupled with two or three-dimensional scatterplots are a common visual for BE detection. But this approach only works if variance correlated with BEs are accounted by the top 2–3 PCs. If BEs are subtle and not correlated with the top 2–3 PCs, then the scatterplots will not work well. Moreover, it is important to note that interpretation of PCA scatterplots does take some skill. Since PCA projects data into orthogonal PCs, the PCs are by construction, independent. This also implies that when constructing scatterplots based on different PCs, sample distributions can appear very different. Moreover, if BEs are correlated with multiple lower PCs, then it is inefficient to plot all PCs to discover batch-correlated variance visually.
The limitations of PCA scatterplots above have spurred search for more convenient and readily interpretable visual aids. Methods like t-Distributed Stochastic Neighbor Embedding (t-SNE) [26] and Uniform Manifold Approximation and Projection (UMAP) [27] embed high-dimensional data into low-dimensional non-linear manifolds, are becoming increasingly popular, especially in the single cell-omics arena. UMAP and t-SNE preserve local structures in data even when converting from high-dimension to low-dimension space. This preservation of local information allows an intuitive visual scan for any BEs in data, absolving the need to scan for the correction BE correlated dimensions. But this does not mean UMAP and t-SNE are perfect. These methods do not preserve global structures well. And so, distances and positions between t-SNE or UMAP clusters are effectively meaningless and not interpretable.
Thus, we feel there is no perfect visualization method, and so, we strongly encourage careful exploration and not to simply rely only on the top 2-3PCs if PCA is used. Or to place undue faith and confidence in UMAP or t-SNE clusters. To further explore data visually for BEs at various levels of granularity, we also recommend use of side-by-side boxplots for visualizing correlations of batch effects with principal components on data [16], and the side-by-side barcharts for investigating gene-gene correlations [7].
Finally, beyond visualizations, statistical methods are useful. For example, we may devise a systematic test for PCs correlated with BEs so that we can identify which orthogonal factors are batch correlated or confounded with class information. This would give us a much better understanding of our data, and also provide us with some targeted mitigation approaches for batch correction [28]. Information across PCs can also be summarized using methods like Principal Variance Component Analysis (PVCA) [29] or guided PCA (gPCA) [30]. Such approaches are complementary with visualizations based on PCA scatterplots, t-SNE and UMAP.
6. Many batch effect correction algorithms use specialized distributions. What about for proteomics?
Recently, we see the rise of dedicated BECAs catered for RNA-seq data (and also, single cell -omics). ComBat-seq is an update of the highly popular ComBat algorithm [31]. ComBat-seq uses a negative binomial regression model that retains the integer nature of count data in RNA-seq, making the batch corrected data compatible with typical RNA-seq differential expression methods that requires integer counts [31]. Changing the background statistical model is highly beneficial --- ComBat-seq corrected data produces improves statistical power while also allowing better control of false positives. And is also very useful in other practical applications such as machine learning data optimization [32]. Like ComBat-seq, svaseq is an update of the popular sva method catering for count data from RNA-seq experiments [11]. Proteomics data, like RNA-seq data, may not follow well theoretical distributions either. We believe BE correction in proteomics would greatly benefit too if proteomic-specific versions of ComBat or SVA were developed, perhaps soon.
Of late, there also seems to be increased interest in one-step BE correction methods (which is prevalent in the single-cell -omics domain [4], [15]). In one-step approaches, batch information is directly accommodated into the analysis. This is in contrast to traditional two-step approaches where data is processed or batch corrected prior to the formal or final analysis (two-step correction) [4]. In gene expression analysis, traditional two-step approaches while more interpretable and can lead towards richer representations of batch information, also induces correlation structures in the BE corrected data, which, if ignored, can produce false positives/negatives during functional analysis [33]. Although one-step approaches seem to be up-and-coming in bulk and single cell RNA-seq, it appears that such integrative one-step approaches are lacking in proteomics. Therefore, we think this is also an interesting development for the future.
7. Post-evaluative: How do I know my batch effect correction was successful?
BE correction while important, is only part of a much larger analytical pipeline. When analyzing high-throughput proteomics data, we are often more interested in the end point, which is to identify interesting proteins that may be causal or important for a particular disease. Therefore, a successful outcome is to retain class information, while minimizing BE confounding.
BE correction is usually deemed successful if batch separation is not observable visually or detectable by PCA methods such as PVCA [29] or gPCA [30]. It is also important to check for retention of class information and how different are the outputs from the BE corrected data against the original. Besides showing batch correlations, gPCA and PVCA can also be tweaked to reveal class information (This is achievable by simply substituting the batch factor with class factor instead). We can also use methods such as root-mean-square deviation (RMSD) or normalized root-mean-square deviation (nRMSD) to evaluate change---this is useful for checking if the data has changed so dramatically post batch processing that we may suspect possible over-correction (or mis-correction).
There is also value in checking how differential proteins changed before and after BE correction. Normally, BE correction will result in both changes in the differential protein list, and their corresponding effect sizes. For differential proteins found in both pre and post batch corrected data (shared), it is useful to note if there is an increment in the effect size, making these proteins more readily detectable (and possibly useful as biomarkers). It is also useful to perform functional analysis based on gene ontology [34], gene sets [35] or biological pathways [36], [37] on the shared proteins, and those which are found only in the pre-batch and post-BE correction complements. If the batch correction is meaningful, we would expect the post-BE complement to have more in common with shared protein functionalities, than with the pre-batch complement. While such analysis are not quantitatively tractable, it does serves as a sanity check.
8. Case study of (early) peptide batch correction
To explore the benefits of early batch correction (albeit in a rather raw manner), we benchmark several pipelines on the BXD Mouse Liver Aging dataset developed by Williams et al. [38] (PXD009160; accessed on November 2021) which was also featureed in Cuklina et als’ proBatch paper [17].
8.1. Dataset
The BXD Mouse Liver Aging Dataset describes a low heterogeneity DIA-SWATH dataset with 2 biological classes (high-fat diet and chow diet) and seven distinct but balanced batches through non-sequential runs. There is also signal drift attributed to a long series of runs. We downloaded the processed peptide matrix (“E1801171630_matrix.xlsx”), which contained minimal missing values at the peptide-level. Drift was corrected by fitting and correcting from a LOESS curve for each batch.
8.2. Batch correction using ComBat
To correct for BEs, we used the Python implementation of ComBat [8], pyCombat [39], available at https://epigenelabs.github.io/pyComBat/. The ComBat algorithm is a location-scale method that assume a Gaussian-like distribution and uses an empirical Bayes method to estimate and correct for additive and multiplicative BEs.
8.3. Analysis pipelines
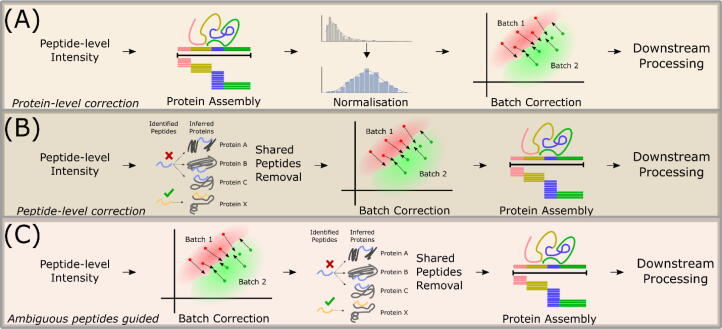
The overall methodology is shown in Fig. 1.We evaluate three scenarios: The first reviews direct BE correction on peptide prior to protein assembly (Fig. 1A). The second is the typical Protein-level BE correction (Fig. 1B). The third scenario depicted in Fig. 1C explores the value of ambiguous peptides in batch correction where we perform peptide-level BE correction where ambiguous peptides are first retained for batch estimation, and then discarded before protein assembly.
Fig. 1.
Summary of all workflows used in this study (A) Protein-level BE correction. (B). (A) Peptide-level BE correction prior to protein assembly. (C) Peptide-level BE correction where ambiguous peptides are first retained for batch estimation, and then discarded before protein assembly.
8.4. Evaluating batch effect correction
For quantifying BEs, we used guided PCA (gPCA) by Reese et al. [30]. Guided PCA uses singular value decomposition to estimate discrete factor associated variance, such as batch or biological class. The method guides the dimension reduction process towards the highest representation of batch variance. Batch associated variance is then derived by comparing guided PCA variance (batch variance, PC1) to unguided PCA variance (total variance, PC1), to derive the delta metric [30].
Our second approach is to perform univariate statistical testing on each principal component (PC). For example, we may run the Kruskal-Wallis (KW) H-test across PC 1 to n for a given data [28], to test correlations with batches and class covariates. This approach allows us to identify which PC was correlated with BEs before and after correction---an effective BE correction approach should demote the PC. Conversely, an effective BE correction should also promote the PC correlation with class effects.
8.5. The impact of early correction is not immediately appreciable
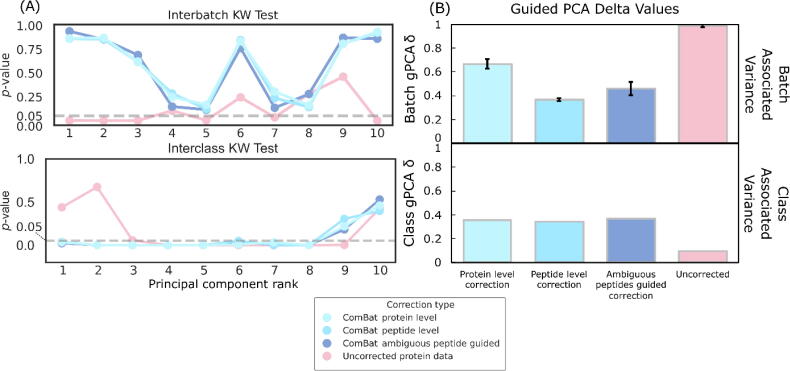
At least on this data, the Kruskal Wallis H-test suggests that protein-level correction and peptide-level correction are highly correlated with each other for both between-classes test and between-batches test (Fig. 2). All scenarios improve class differentiation while demoting batch effects when evaluated against uncorrected data. None of the seemingly sensible strategies e.g., retaining ambiguous peptides for batch estimation or early correction on methods demonstrate clear superiority.
Fig. 2.
Evaluating BE correction across two methods. (A) A line plot showing the relationship between PCs and associative p-values based on Kruskal-Wallis test. The top and bottom panels shows associations with batch effects and class effects respectively. (B) Barcharts of guided PCA (gPCA) delta values for batch (top) and class (bottom) effects. For both (A) and (B), an ideal method is one that promotes class effects while demoting batch effects.
8.6. Case study verdict
In this case study, despite the attractiveness and apparent logic of alternative batch correction scenarios, we do not see any advantage. This is not to say these other scenarios do not work universally. Perhaps, there are specific use cases and scenarios where some scenarios could manifest superiority to others. However, this warrants deeper investigation, and development of gold-standard scenarios so that we may better interpret the results. This is also valuable, as it will help the community determine the best course of BE correction given their data.
9. Conclusion
BE correction is a highly complex data processing step, important for proteomic analysis. Despite the advent of better data acquisition technologies, BE correction problems persist. There is no single best way for performing BE correction---it is therefore important to understand the nature of the dataset, while also keeping in mind how different normalization methods and BE correction methods affect each other. Finally, while ideas such as early batch correction seems attractive and sensible, it did not work well in our case study and may not apply to every dataset.
CRediT authorship contribution statement
Ser-Xian Phua: Methodology, Visualization. Kai-Peng Lim: Methodology, Visualization. Wilson Wen-Bin Goh: Conceptualization, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
WWBG acknowledge support from an MOE AcRF Tier 1 award (RG35/20).
Author contributions
SXP and KPL implemented the case study. WWBG supervised, edited and wrote the manuscript.
References
- 1.Goh W.W.B., Wong L. Dealing with confounders in omics analysis. Trends Biotechnol. 2018;36(5):488–498. doi: 10.1016/j.tibtech.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Nygaard V., Rødland E.A., Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17(1):29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L., Chi-Hau Sue A., Bin Goh W.W. Examining the practical limits of batch effect-correction algorithms: When should you care about batch effects? J Genetics Genomics. 2019;46(9):433–443. doi: 10.1016/j.jgg.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Goh W.W.B., Yong C.H., Wong L. Are batch effects still relevant in the age of big data? Trends Biotechnol. 2022 doi: 10.1016/j.tibtech.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Käll L., Vitek O. Computational mass spectrometry-based proteomics. PLoS Comput Biol. 2011;7(12):e1002277. doi: 10.1371/journal.pcbi.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang T., Wang J., Yu W., He Z. Protein inference: a review. Briefings Bioinf. 2012;13(5):586–614. doi: 10.1093/bib/bbs004. [DOI] [PubMed] [Google Scholar]
- 7.Leek J.T., et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 9.Oytam Y., Sobhanmanesh F., Duesing K., Bowden J.C., Osmond-McLeod M., Ross J. Risk-conscious correction of batch effects: maximising information extraction from high-throughput genomic datasets. BMC Bioinf. 2016;17(1):332. doi: 10.1186/s12859-016-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3(9):e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leek J.T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucl Acids Res. 2014;42(21):e161–e. doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe A.E., et al. Practical impacts of genomic data ‘cleaning’ on biological discovery using surrogate variable analysis. BMC Bioinf. 2015;16(1):372. doi: 10.1186/s12859-015-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papiez A., Marczyk M., Polanska J., Polanski A. BatchI: Batch effect Identification in high-throughput screening data using a dynamic programming algorithm. Bioinformatics. 2019;35(11):1885–1892. doi: 10.1093/bioinformatics/bty900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J., et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J. 2010;10(4):278–291. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran H.T.N., et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21(1):12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh W.W.B., Wong L. Protein complex-based analysis is resistant to the obfuscating consequences of batch effects –- a case study in clinical proteomics. BMC Genomics. 2017;18(S2):142. doi: 10.1186/s12864-017-3490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Čuklina J., et al. Diagnostics and correction of batch effects in large-scale proteomic studies: a tutorial. Mol Syst Biol. 2021;17(8):Aug. doi: 10.15252/msb.202110240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belorkar A., Wong L. GFS: fuzzy preprocessing for effective gene expression analysis. BMC Bioinf. 2016;17(S17):540. doi: 10.1186/s12859-016-1327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Lee J., Goh W.W.B. An investigation of how normalisation and local modelling techniques confound machine learning performance in a mental health study. Heliyon. 2022;8(5):e09502. doi: 10.1016/j.heliyon.2022.e09502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giuliani A. The application of principal component analysis to drug discovery and biomedical data. Drug Discovery Today. 2017;22(7):1069–1076. doi: 10.1016/j.drudis.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Giuliani A., Colosimo A., Benigni R., Zbilut J.P. On the constructive role of noise in spatial systems. Phys Lett A. 1998;247(1–2):47–52. doi: 10.1016/S0375-9601(98)00570-2. [DOI] [Google Scholar]
- 22.Goh W.W.B., Wang W., Wong L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017;35(6):498–507. doi: 10.1016/j.tibtech.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Graw S., et al. proteiNorm – A user-friendly tool for normalization and analysis of TMT and label-free protein quantification. ACS Omega. 2020;5(40):25625–25633. doi: 10.1021/acsomega.0c02564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenes A., Hukelmann J., Bensaddek D., Lamond A.I. Multibatch TMT reveals false positives, batch effects and missing values. Mol Cell Proteomics. 2019;18(10):1967–1980. doi: 10.1074/mcp.RA119.001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun P.Y.Q., Goh W.W.B. Why batch sensitization is important for missing value imputation. Research Square. 2022 doi: 10.21203/rs.3.rs-1328989/v1. [DOI] [Google Scholar]
- 26.van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learning Res. 2008;9:2579–2605. [Google Scholar]
- 27.McInnes L, Healy J, Melville J, UMAP: uniform manifold approximation and projection for dimension reduction, 2018, doi: 10.48550/ARXIV.1802.03426.
- 28.Goh W.W.B., Sng J.-C.-G., Yee J.Y., Lee T.-S., Wong L., Lee J. Can peripheral blood-derived gene expressions characterize individuals at ultra-high risk for psychosis? Comput Psychiatry. 2017;1:168. doi: 10.1162/CPSY_a_00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Bushel P.R., Chu T.-M., Wolfinger R.D. In: Batch effects and noise in microarray experiments. Scherer A., editor. John Wiley & Sons Ltd; Chichester, UK: 2009. Principal variance components analysis: estimating batch effects in microarray gene expression data; pp. 141–154. [DOI] [Google Scholar]
- 30.Reese S.E., et al. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics. 2013;29(22):2877–2883. doi: 10.1093/bioinformatics/btt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Parmigiani G, Johnson WE, ComBat-seq: batch effect adjustment for RNA-seq count data, NAR Genomics and Bioinformatics, 2(3), p. lqaa078, 2020, doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed]
- 32.Wang LR, Choy XY, Bin Goh WW, Doppelgänger Spotting in Biomedical Gene Expression Data, iScience, p. 104788, 2022, doi: 10.1016/j.isci.2022.104788. [DOI] [PMC free article] [PubMed]
- 33.Li T, Zhang Y, Patil P, Johnson WE, “Overcoming the impacts of two-step batch effect correction on gene expression estimation and inference,” Biostatistics, p. kxab039, Dec. 2021, doi: 10.1093/biostatistics/kxab039. [DOI] [PMC free article] [PubMed]
- 34.Zheng Q, Wang X-J, “GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis,” Nucleic Acids Research, vol. 36, no. suppl_2, pp. W358–W363, Jul. 2008, doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed]
- 35.Zyla J., Marczyk M., Domaszewska T., Kaufmann S.H.E., Polanska J., Weiner J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics. 2019;35(24):5146–5154. doi: 10.1093/bioinformatics/btz447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutmon M., Lotia S., Evelo C.T., Pico A.R. WikiPathways App for Cytoscape: making biological pathways amenable to network analysis and visualization. F1000Res. 2014;3:152. doi: 10.12688/f1000research.4254.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nersisyan L, Samsonyan R, Arakelyan A, “CyKEGGParser: tailoring KEGG pathways to fit into systems biology analysis workflows,” F1000Res, vol. 3, p. 145, Aug. 2014, doi: 10.12688/f1000research.4410.2. [DOI] [PMC free article] [PubMed]
- 38.Williams EG, et al., Multiomic profiling of the liver across diets and age in a diverse mouse population, Cell Systems, p. S2405471221003446, Oct. 2021, doi: 10.1016/j.cels.2021.09.005. [DOI] [PMC free article] [PubMed]
- 39.Behdenna A., Haziza J., Azencott C.-A., Nordor A. pyComBat, a Python tool for batch effects correction in high-throughput molecular data using empirical Bayes methods. bioRxiv. 2020 doi: 10.1101/2020.03.17.995431. [DOI] [PMC free article] [PubMed] [Google Scholar]