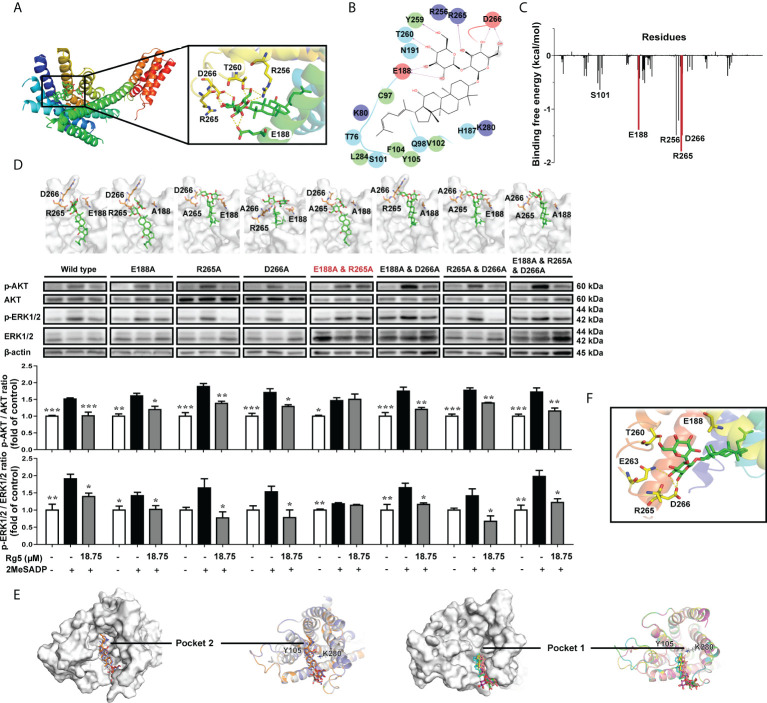
Figure 3.
Interaction sites of Rg5 with P2RY12. (A) P2RY12 and Rg5 complex structure, and binding pocket with Rg5. (B) Two-dimensional interaction map of interactions between Rg5 and P2RY12. (C) Energy decomposition of amino acid residues in P2RY12 and Rg5 based on MM/PBSA free energy calculation. (D) Structures of P2RY12 mutation variants with Rg5 bound were shown in the upper panel, and Western blot analysis in the lower panel displayed the protein expression and phosphorylation levels of AKT (n = 4) and ERK (n = 4). All the data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. 2MesADP group. (E) Hypothetical binding modes of Rg5 to P2RY12 mutation variants obtained by molecular docking. The binding conformation of Rg5 in each P2RY12 mutant were colored as follows: white, original; slate, E188A; orange, E188A and R265A; magenta, R165A; pink, D266A; cyan, E188A and D266A; yellow, R265A and D266A; green, E188A and R265A and D266A. (F) A view of the P2RY12 mutant/Rg5 complex.