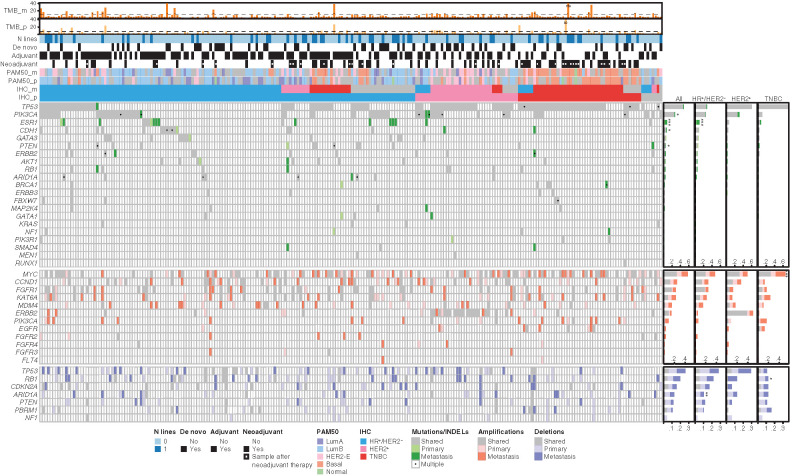
Figure 2.
Repertoire of somatic gene alterations. Oncoplot of the relevant genomic alterations in the set of 242 patients with available Target Gene Sequencing (TGS) data for primary and metastatic samples. From top to bottom, the oncoplot includes three sections: tumor mutational burden (TMB), clinical data, and genomic alterations. TMB section shows the bar plots of TMB in primary and metastatic samples. Dashed lines refer to the TMB threshold used to define high-TMB patients based on the 90th percentile of the TMB distribution (corresponding to 8 for primary and 11 for metastatic samples). Clinical data section includes information about the number of treatment lines for metastatic disease, de novo metastatic disease, adjuvant and neoadjuvant therapy, and molecular subtype information in primary and metastatic samples by PAM50 and IHC. Genomic alterations are classified as shared (if present in both primary and metastatic samples), primary (private to primary sample), and metastatic (private to metastatic samples). Genomic alterations include driver mutations (single-nucleotide variants and insertions/deletions) in driver genes, amplifications in oncogenes, and deletions in tumor suppressor genes. On the right, the bar plots summarize, for each gene, the frequency of shared, private to primary, and private to metastatic events. The asterisks refer to genes showing significant difference in terms of alteration frequency in metastatic compared to primary samples (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Genes (SNVs) with significant positive selection on missense mutations and/or truncating substitutions on the dN/dS analysis are represented in the figure. We have included in the figure CNVs of genes known as breast cancer drivers.