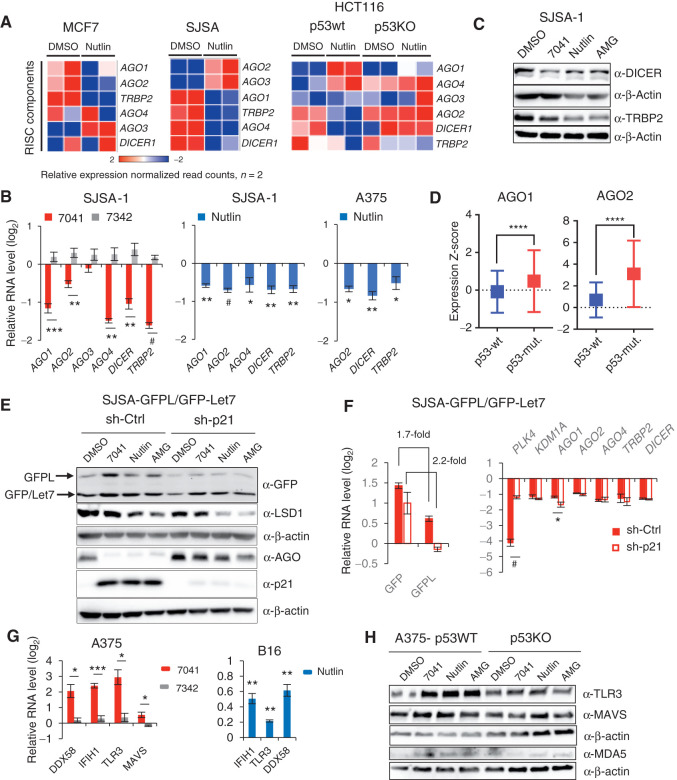
Figure 3.
Inhibition of dsRNA processing genes and RNA-silencing complex (RISC) activity and induction of dsRNA sensors upon p53 activation. A, Heat maps demonstrating downregulation of RISC genes upon p53 activation by nutlin in three cancer cell lines, as assessed by RNA-seq. B, Immunoblot analysis depicts downregulation of DICER and TRBP2 after p53 activation by 7041, nutlin, or AMG-232 in SJSA-1 cells. C, Genes encoding RISC components are repressed after p53 activation by 7041 (red bars) and nutlin (blue bars) in SJSA-1 and A375 cells, as assessed by qPCR (n = 3). D, Box plots depict higher levels of expression of AGO1 and AGO2 genes in breast tumors harboring mutant TP53 compared with wild-type ones. RNA-seq data are from TCGA. Relative expression values are presented as mRNA expression Z-scores (Mann–Whitney test; ****, P ≤ 0.0001). E and F, Inhibition of RISC activity upon p53 activation by nutlin, 7041, and AMG-232 is largely p21-independent. The expression of GFPL and GFP was measured by immunoblot (E) and real-time qPCR (F) in SJSA-1 cells stably expressing dual reporters GFPL/GFP-let-7 and transduced with scrambled shRNA or p21 shRNA. E, Immunoblotting of LSD1 and AGO proteins (detected by pan-AGO antibody) upon p53 activation in the presence or absence of p21. F, p21-independent repression of RISC genes after p53 reactivation by 7041 (red bars), as assessed by qPCR (n = 3). G, dsRNA receptors are induced upon p53 activation by 7041 (red bars) and nutlin (blue bars) in A375 and B16 cells, as assessed by qPCR, n = 3. H, Induction of TLR3, MDA5, and MAVS in a p53-dependent manner in A375 cells after p53 activation by three MDM2 inhibitors, as detected by immunoblot. β-Actin was used as loading control. B, F, G, Student t tests. Error bars, SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0,0001, n = 3.