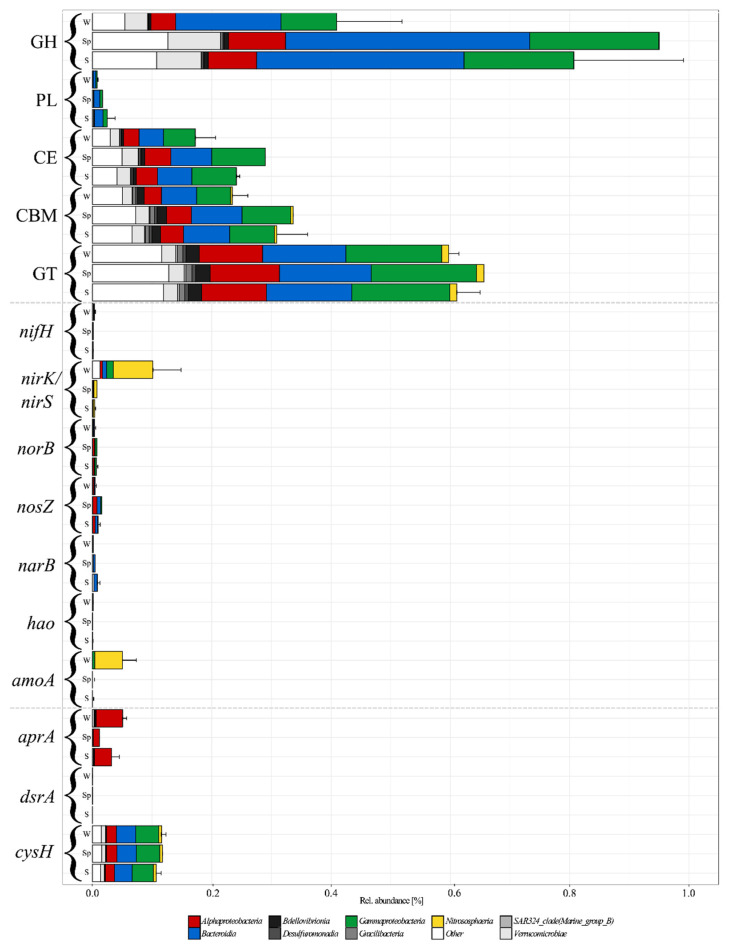
Figure 7.
Rel. abundance of predicted CAZyme classes (glycoside hydrolases (GH), polysaccharide lyases (PL), carbohydrate esterases (CE), carbohydrate-binding modules (CBM), and glycosyl transferase (GT)) and marker gene abundances for the three distinct seasons (Winter (W), Spring (Sp), and Summer (S)). The rel. abundance is shown as % of total genes detected in the PiCRUST2 inferred metagenome for each sample. The error bars show the variability in the four stations pooled for the winter sample and the two stations for the summer sample. No error bar is available for spring since only one sample was taken in this season. The bars include the predicted taxonomic assignment of the respective CAZyme classes and marker genes on class level.