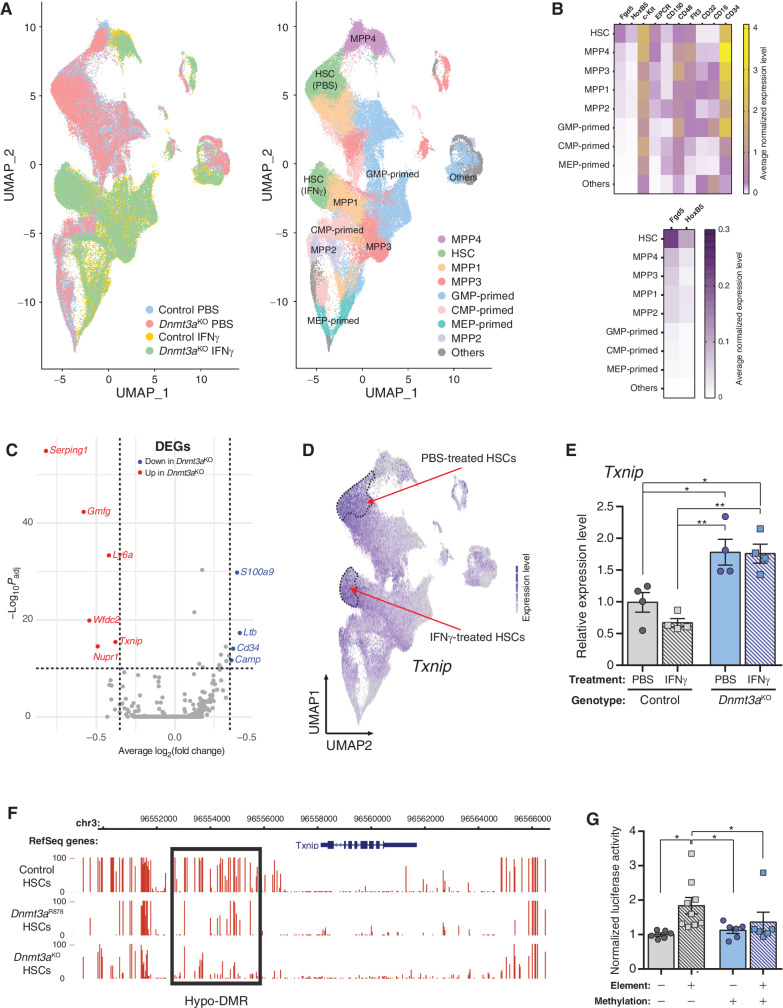
Figure 5.
Txnip is a DNA methylation–sensitive gene involved in HSC stress response. A, UMAP clustering of single-cell RNA-seq data of control and Dnmt3aKO Lineage− c-Kit+EPCR+ hematopoietic cells after treatment with PBS or IFNγ. Left, clustering of cells by genotype and treatment. Right, assignment of cell identities to cell clusters. Two clusters of HSCs are identified, which represents HSCs from both genotypes separated by treatment. B, Marker gene expression used to assign identities to cell clusters. C, Volcano plot of differentially expressed genes (DEG) between IFNγ-treated control and Dnmt3aKO HSCs. D,Txnip expression in UMAP-clustered scRNA-seq data. Dashed lines outline HSC clusters. E,Txnip expression in HSCs purified from mice treated with PBS or IFNγ (n = 4). F, DNA methylation profile of Txnip locus in HSCs determined by WGBS. Height of red bar indicates methylation level of individual CpG. Black box denotes hypomethylated DMR in Dnmt3a-mutant HSCs. G, Luciferase activity of Txnip regulatory element in methylation-sensitive reporter assay. Firefly luciferase activity was measured 48 hours posttransfection and normalized to Renilla luciferase levels (n = 6–9). One-way ANOVA; *, P < 0.05; **, P < 0.01. Data represent mean ± SEM.