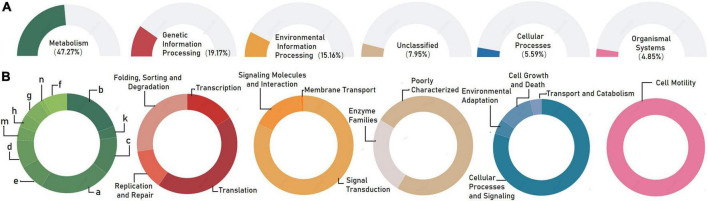
FIGURE 3.
Functional prediction of the whole bacterial microbiome associated with Karlodinium veneficum in KEGG categories at level 1 (A) and level 2 (B). Gene functions were predicted from 16S rRNA gene-based microbial compositions using the PICRUSt algorithm to make inferences from KEGG annotated databases. Relative signal intensity was normalized by the number of the genes for each indicated metabolic pathway. a: amino acid metabolism; b: carbohydrate metabolism; c: energy metabolism; d: metabolism of cofactors and vitamins; e: lipid metabolism; f: nucleotide metabolism; g: xenobiotics biodegradation and metabolism; h: glycan biosynthesis and metabolism; m: metabolism of terpenoids and polyketides; n: biosynthesis of other secondary metabolites.