Figure 1. Some examples of recent structures that have been determined by cryo-EM.
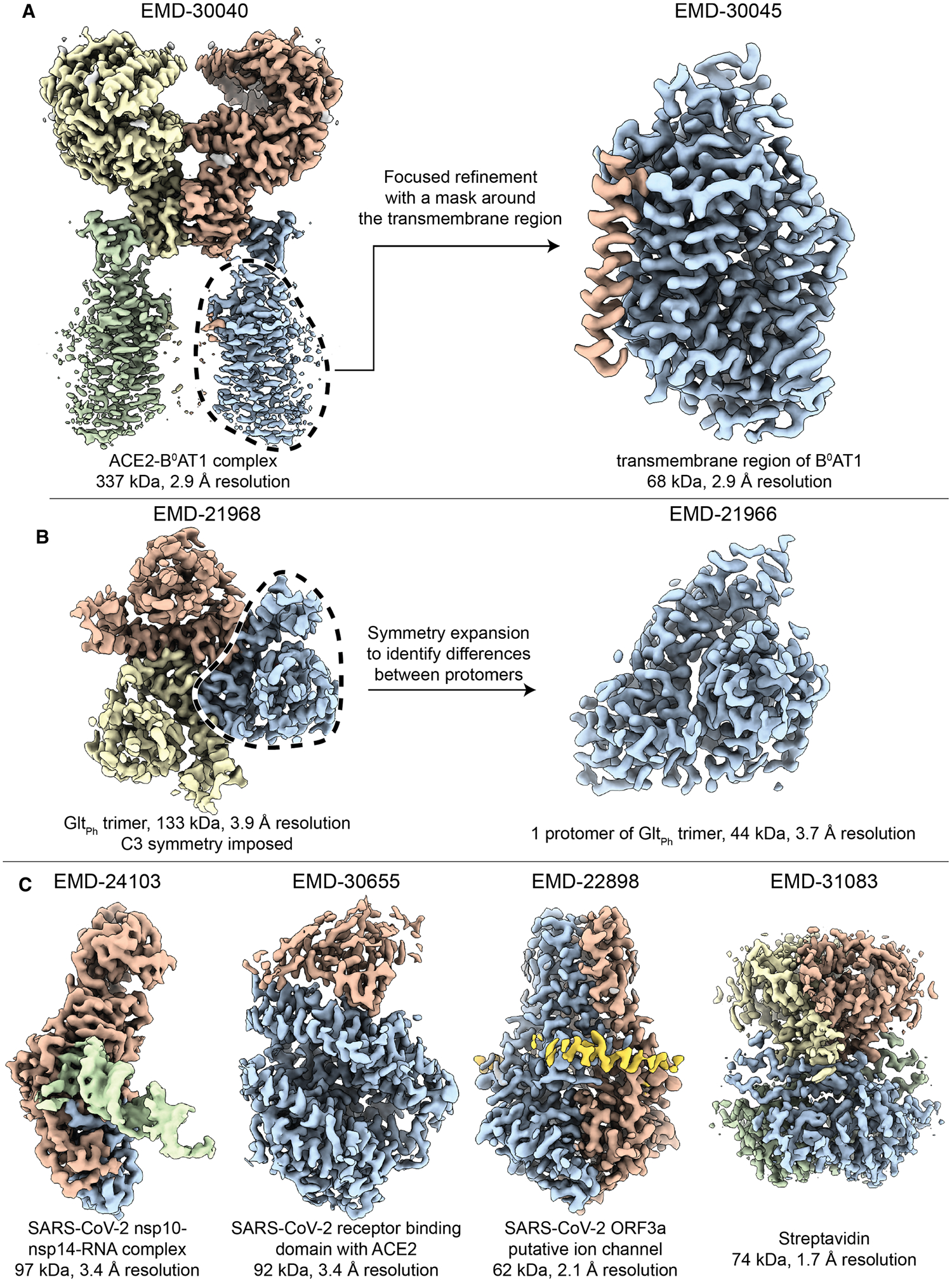
Accession numbers for each of these illustrations are indicated above the respective maps shown here. (A) The two-fold symmetric structure of the angiotensin-converting enzyme 2 (ACE2) bound to the membrane protein B0AT1, was determined at a reported resolution of better than 3 Å [73]. However, the transmembrane regions of B0AT1 were not well-resolved in this map. The mask-and-align approach was used to substantially improve the resolution of this 68 kDa region. (B) The three-fold symmetric structure of the archaeal glutamate transporter GltPh was determined with a reported resolution of 3.9 Å resolution. A specialized type of mask-and-align approach, which enables asymmetric units within a symmetric complex to be structurally analyzed in isolation (reviewed in [74]), was used to reveal the presence of distinct conformers among the protomers [75]. (C) Four recent single particle reconstructions of <100 kDa complexes at increasing resolution. Shown from left to right are: The SARS-CoV-2 exoribonuclease (nsp10–nsp14) bound to RNA [17], the pangolin coronavirus receptor binding domain bound to human ACE2 [19], the SARS-CoV-2 open reading frame 3a putative ion channel [22], and streptavidin from Streptomyces avidinii acquired using a Krios G4 electron microscope equipped with a cold FEG, Selectris energy filter and Falcon IV direct detector (personal communication with Dr. Tomohiro Nishizawa).
