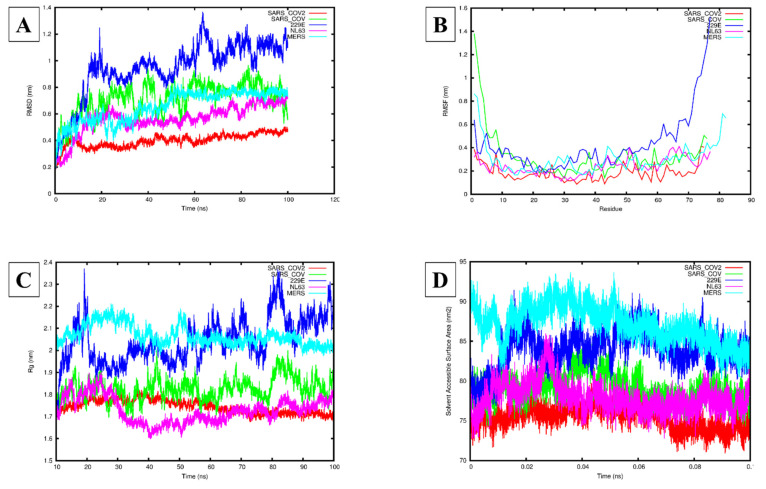
Figure 6.
Molecular dynamics simulations of the human (h) coronavirus (CoV) envelope (E) proteins for SARS-CoV-1, -2, MERS-CoV, HCoV-229E, and HCoV-NL63. (A) The root mean square deviation (RMSD) (nm) of each of the modelled hCoV E proteins over 100 ns. (B) The root mean square fluctuation (RMSF) (nm) of each residue for the modelled hCoV E proteins. (C) The radius of gyration (Rg) (nm) of each of the modelled hCoV E proteins over 100 ns. (D) The solvent-accessible surface area (SASA) (nm2) of each of the modelled hCoV E proteins over 0.1 ns. Red: SARS-CoV-2 E protein, Green: SARS-CoV-1 E protein, Blue: HCoV-229E E protein, Pink: HCoV-NL63 E protein, Cyan: MERS-CoV E protein.