Fig. 1: HCV infection leads to genome-wide loss of DNA methylation and locus-specific hypermethylation.
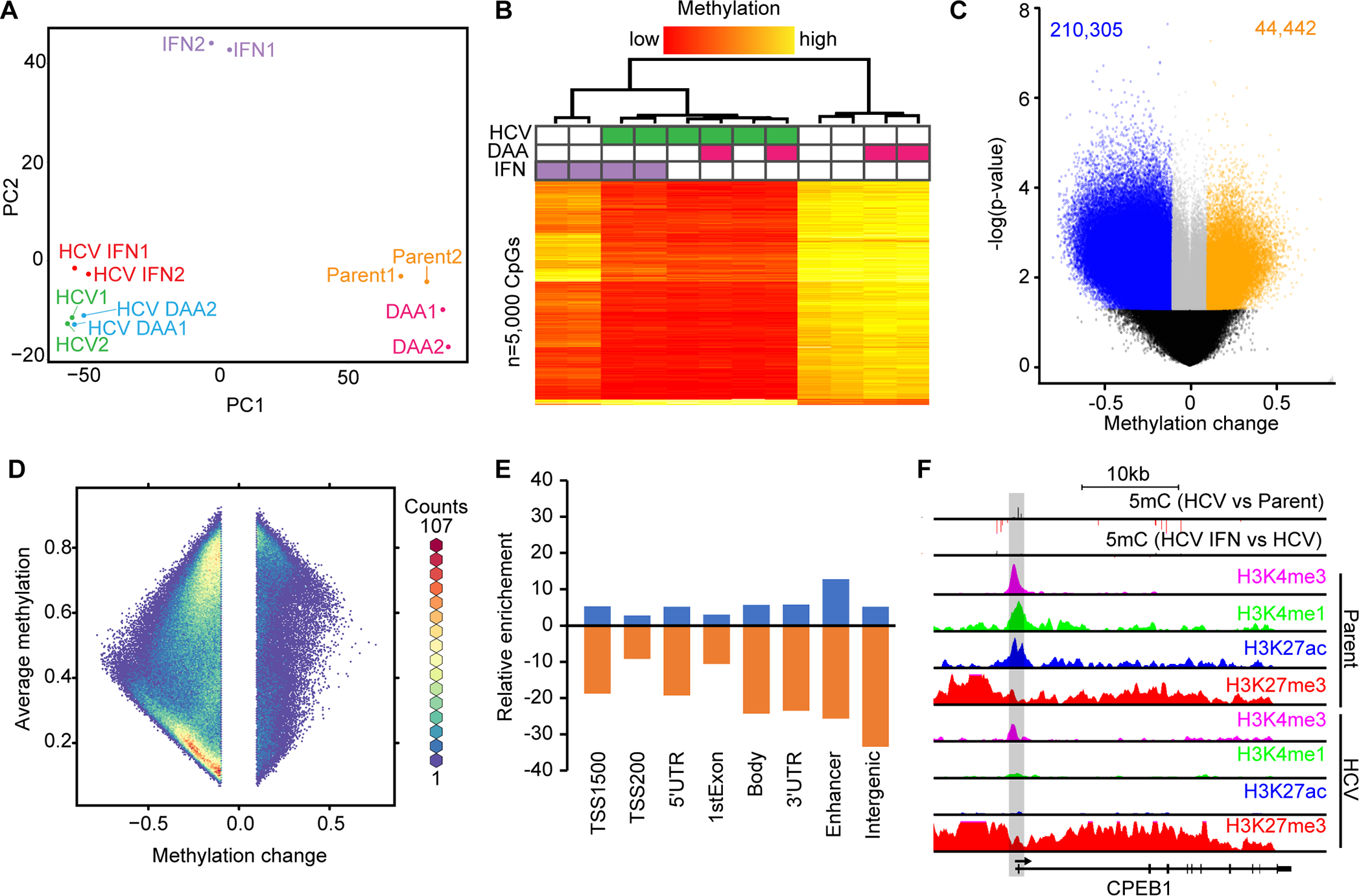
(A) PCA of Hu1545 and its derivatives used in this study. Each condition has two biological replicates. (B) Unsupervised hierarchical clustering of the 5,000 most variable CpGs across the six biological conditions indicated (in duplicate). A color bar is shown. (C) A volcano plot of DNA methylation changes against significance in HCV-infected Hu1545 cells compared to parent. Statistically significant hypermethylation (orange) and hypomethylation (blue) events are shown. (D) A hexbin plot of changes in methylation in HCV infected cells against the average methylation of all samples (Hu1545 Parent + HCV). (E) Bar plot depicting the distribution of HCV-induced DNA methylation changes across features on the 850k array. Enhancers are regions marked by both H3K4me1 and H3K27ac. (F) UCSC browser view of a representative locus (CPEB1) demonstrating promoter hypermethylation in HCV-infected cells. The second track depicts methylation changes induced by interferon treatment in HCV-infected cells. Also shown are histone modifications by ChIP-seq for H3K4me3, H3K27ac, H3K4me1, and H3K27me3 in parent (top four) and HCV (bottom four) Hu1545 cells.
