Abstract
Background
Direct evaluation of vascular inflammation in patients with COVID-19 would facilitate more efficient trials of new treatments and identify patients at risk of long-term complications who might respond to treatment. We aimed to develop a novel artificial intelligence (AI)-assisted image analysis platform that quantifies cytokine-driven vascular inflammation from routine CT angiograms, and sought to validate its prognostic value in COVID-19.
Methods
For this prospective outcomes validation study, we developed a radiotranscriptomic platform that uses RNA sequencing data from human internal mammary artery biopsies to develop novel radiomic signatures of vascular inflammation from CT angiography images. We then used this platform to train a radiotranscriptomic signature (C19-RS), derived from the perivascular space around the aorta and the internal mammary artery, to best describe cytokine-driven vascular inflammation. The prognostic value of C19-RS was validated externally in 435 patients (331 from study arm 3 and 104 from study arm 4) admitted to hospital with or without COVID-19, undergoing clinically indicated pulmonary CT angiography, in three UK National Health Service (NHS) trusts (Oxford, Leicester, and Bath). We evaluated the diagnostic and prognostic value of C19-RS for death in hospital due to COVID-19, did sensitivity analyses based on dexamethasone treatment, and investigated the correlation of C19-RS with systemic transcriptomic changes.
Findings
Patients with COVID-19 had higher C19-RS than those without (adjusted odds ratio [OR] 2·97 [95% CI 1·43–6·27], p=0·0038), and those infected with the B.1.1.7 (alpha) SARS-CoV-2 variant had higher C19-RS values than those infected with the wild-type SARS-CoV-2 variant (adjusted OR 1·89 [95% CI 1·17–3·20] per SD, p=0·012). C19-RS had prognostic value for in-hospital mortality in COVID-19 in two testing cohorts (high [≥6·99] vs low [<6·99] C19-RS; hazard ratio [HR] 3·31 [95% CI 1·49–7·33], p=0·0033; and 2·58 [1·10–6·05], p=0·028), adjusted for clinical factors, biochemical biomarkers of inflammation and myocardial injury, and technical parameters. The adjusted HR for in-hospital mortality was 8·24 (95% CI 2·16–31·36, p=0·0019) in patients who received no dexamethasone treatment, but 2·27 (0·69–7·55, p=0·18) in those who received dexamethasone after the scan, suggesting that vascular inflammation might have been a therapeutic target of dexamethasone in COVID-19. Finally, C19-RS was strongly associated (r=0·61, p=0·00031) with a whole blood transcriptional module representing dysregulation of coagulation and platelet aggregation pathways.
Interpretation
Radiotranscriptomic analysis of CT angiography scans introduces a potentially powerful new platform for the development of non-invasive imaging biomarkers. Application of this platform in routine CT pulmonary angiography scans done in patients with COVID-19 produced the radiotranscriptomic signature C19-RS, a marker of cytokine-driven inflammation driving systemic activation of coagulation and responsible for adverse clinical outcomes, which predicts in-hospital mortality and might allow targeted therapy.
Funding
Engineering and Physical Sciences Research Council, British Heart Foundation, Oxford BHF Centre of Research Excellence, Innovate UK, NIHR Oxford Biomedical Research Centre, Wellcome Trust, Onassis Foundation.
Introduction
Cardiovascular complications have emerged as a key feature of COVID-19 and drive adverse outcomes.1 Inflammation-induced injury of the vascular endothelium is crucial in COVID-19 pathogenesis because it seems to drive a procoagulant state that contributes to multiorgan failure.2, 3
Direct evaluation of vascular inflammation in COVID-19 would pave the way for more efficient trials of new treatments, identify patients who might be at risk of long-term complications, and help detect responders to high-risk treatments, such as steroids.4
Research in context.
Evidence before this study
The field of radiotranscriptomics (ie, the combination of transcriptomics with radiomics) in CT imaging is still in its infancy. Radiomics have been previously applied, mainly in cancer research, to provide prognostic information for a series of morbidities. In COVID-19, previous studies have investigated lung-based radiomics or deep learning modelling based on CT imaging to predict disease status. We searched PubMed from Jan 1, 2010, to March 19, 2022, for full-text articles investigating prognosis of COVID-19 published in any language using the search algorithm (“COVID-19” OR “SARS-CoV-2”) AND (“computed tomography”) AND (prognosis[MeSH Terms]) AND (“inflammation”) AND (“radiomics”). We found one machine learning model combining lung radiomics with clinical information by Shiri and colleagues, which was shown to predict in-hospital death from COVID-19 in a test set of 46 patients, while one further study found that a positive PET scan indicated increased risk of symptomatic SARS-CoV-2 infection and admission to hospital in 13 patients with COVID-19. However, we found no study that focused on non-invasive assessment of vascular inflammation in COVID-19, or that combined transcriptomic data with radiomics to quantify vascular inflammation in these patients, indicating the necessity of the present study.
Added value of this study
This study presents the first radiotranscriptomic platform that allows development of imaging signatures of vascular inflammation from routine CT angiograms. This platform can be used to quantify the degree of vascular inflammation in conditions such as COVID-19 (cytokine-driven vascular inflammation), and can help us stratify the severity of new upcoming variants; it could also potentially guide the deployment of specific anti-inflammatory or even anti-thrombotic treatments to those patients who need them. Additionally, this platform could be used to construct radiotranscriptomic signatures of different types of vascular inflammation, and could be particularly useful in pharmaceutical drug development pipelines.
Implications of all the available evidence
The new radiotranscriptomic method described here allows training of digital fingerprints of vascular disease, enabling the performance of digital vascular biopsies from routine CT angiograms. To the best of our knowledge, this is the first application of this method in COVID-19, which now allows us to detect those patients who have cytokine-driven vascular inflammation—the subgroup of patients with COVID-19 who are at risk of thrombosis and high mortality. This subgroup of patients can benefit from systemic corticosteroid treatment with dexamethasone. This method could, therefore, allow clinicians to determine who will benefit from systemic corticosteroid treatment at an early timepoint, allowing targeted prescribing and avoidance of complications from unnecessary administration of dexamethasone to non-responders.
Perivascular adipose tissue (PVAT) has a wide range of bidirectional interactions with the vascular wall.5 Recent evidence suggests that PVAT senses inflammatory signals coming from the arterial wall, activates perivascular lipolysis, and supresses adipogenesis, resulting in accumulation of small and fat-free adipocytes in PVAT surrounding inflamed arteries.6 This response is also accompanied by perivascular oedema, due to the increased permeability of the inflamed perivascular arterioles,7 whereas different types of inflammation might lead to additional perivascular angiogenesis and fibrosis.8, 9 These vessel-driven changes in the histological texture of PVAT can be used to build molecular fingerprints that characterise and quantify the extent of vascular inflammation.
We recently described a method that uses radiomic analysis of CT images to characterise changes in the texture of PVAT driven by inflammatory signals originating from the adjacent arterial wall.10 By applying quantitative radiotranscriptomics, radiomic signatures of PVAT can be developed and trained against the transcriptomic profile of arterial tissue biopsies.9 We developed a radiotranscriptomic platform that linked RNA sequencing data from human internal mammary arteries to radiomic data of the perivascular space, extracted from CT angiography images. This platform was used to train a radiotranscriptomic signature in the PVAT around the internal mammary arteries, to best describe the degree of cytokine-driven arterial inflammation. We then tested the performance of this signature as a biomarker of vascular inflammation in COVID-19, and explored its prognostic value for in-hospital hard endpoints.
Methods
Study design
A comprehensive study workflow diagram for this prospective outcomes validation study is provided in the appendix (p 17) and further information on recruitment is also given in the appendix (pp 1, 18). This study was approved by the local research ethics committee of the University of Oxford, Oxford, UK (Oxford REC C 11/SC/0140). Detailed information on ethical approval and patient consent for the populations in the multiple study arms are given in the appendix (p 1). The study complied with the Declaration of Helsinki.
Study arm 1 (design of the novel radiotranscriptomic imaging signature, C19-RS) comprised 55 patients undergoing cardiac surgery from the Oxford Heart, Vessels and Fat (Ox-HVF) cohort. Segments of internal mammary arteries were collected for transcriptomic analysis, and the RNA sequencing data were linked with coronary CT angiogram (CCTA) imaging done prospectively after patients provided written informed consent, and used as ground truth for machine learning modelling, to train a radiotranscriptomic signature of cytokine-driven inflammation (C19-RS), as described below.
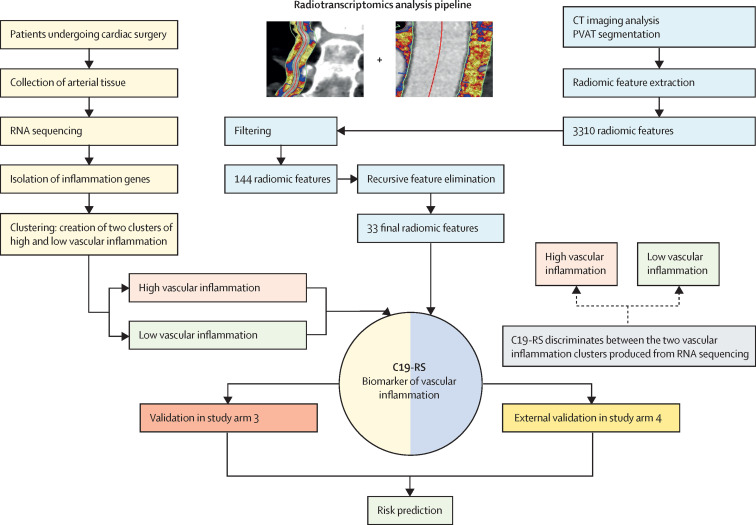
In study arm 1, we created a platform for development of novel imaging biomarkers of vascular inflammation, by applying quantitative radiotranscriptomics to images from standard CT angiography. We used RNA sequencing data of human internal mammary arteries to build a transcriptomic fingerprint of cytokine-driven inflammation in human arteries, similar to what is observed in COVID-19. We then used that transcriptomic fingerprint as the ground truth, and applied machine learning to train a radiomic signature of the perivascular space around the internal mammary artery and the aorta of the same patients (extracted from research coronary CT angiograms of the same patients) to best match that transcriptomic fingerprint of vascular cytokine-driven inflammation (C19-RS). A detailed workflow of the steps involved in the radiotranscriptomic platform is presented in figure 1 .
Figure 1.
Workflow for building the radiotranscriptomic signature C19-RS
Workflow depicting the multiple steps taken to develop the radiomic signature C19-RS. To limit our analysis to radiomic features that could be of value as imaging biomarkers, we did a series of filtering steps, to exclude features that are not stable in test–retest analyses, features that are highly correlated with each other, and features that are significantly correlated with BMI or intrathoracic adipose tissue volume, to retain only features that predict the outcome variable with the same direction within study arm 1 and 20% of the exploratory study arm 3 subpopulation. Finally, recursive feature elimination with a random forest algorithm and repeated five-times cross-validation showed a plateau in the accuracy of the trained model with 33 final features. Those features were next used within the study arm 1 population to train an XGBoost algorithm using decisions trees in order to identify patients with activated inflammatory pathways within their arterial vasculature. The raw product of the algorithm was named C19-RS. PVAT=perivascular adipose tissue.
We have previously shown that inflammation in the internal mammary artery is a suitable surrogate for the inflammatory status of the entire vasculature since it is not affected by atherosclerotic plaque.11, 12 Therefore, the transcriptomic profile of the internal mammary artery provides the optimal ground truth in a machine learning exercise to build a CT-radiomic signature of cytokine-induced arterial inflammation from its PVAT. The internal mammary artery also offers a unique arterial conduit for radiotranscriptomic analysis, given its accessibility during cardiac surgery and the fact that it provides homogeneous PVAT along its length, enabling reliable extraction of radiomic features from its perivascular space, by analysing CCTA and CT pulmonary angiography (CTPA) images.
Study arm 2 (testing the ability of C19-RS to change in patients with COVID-19) comprised 44 patients from the Oxford University Hospitals National Health Service (NHS) Foundation Trust who underwent CCTA imaging before the COVID-19 pandemic and were recruited prospectively. 22 of these patients who developed COVID-19 underwent repeat CCTA within 6 months of SARS-CoV-2 infection, and 22 patients who had no history of SARS-CoV-2 infection at the time of screening (controls with pre-pandemic paired scans, matched for age, sex, and BMI) also underwent repeat CCTA (appendix p 36), as described in the appendix (pp 5–6). This was a substudy of the Oxford Risk Factors and non-invasive Imaging (ORFAN) study (ClinicalTrials.gov, NCT05169333), which received national flagship status by the National Institute of Health Research and the British Heart Foundation (The UK COVID-19 Cardiovascular Research Consortium).
Study arm 3 (validation of the ability of C19-RS to predict in-hospital mortality in COVID-19, and its interaction with anti-inflammatory treatments) comprised 418 patients from the Oxford University Hospitals NHS Foundation Trust undergoing CTPA, who were treated in hospital between March 13, 2020, and Jan 7, 2021, during the first and second waves of the pandemic (appendix p 18). 284 individuals were verified as SARS-CoV-2 positive, based on either a positive RT-PCR test on a pharyngeal swab or the presence of clinicoradiological features on CT imaging. The remaining patients served as controls and were admitted to hospital for other reasons and had a clinically indicated CTPA. The study arm 3 population that was identified during the first UK wave (March–June, 2020; n=269), and before the introduction of routine dexamethasone treatment that could confound the study results, was split randomly into a 20% exploratory subpopulation and an 80% validation subpopulation. The exploratory subpopulation was used for radiomic feature filtering as outlined in the appendix (pp 10, 17). The remaining 80% validation subpopulation pooled together with the rest of the study arm 3 population (appendix p 17; n=331) was used to test C19-RS. Viral whole-genome sequencing was done in patients in study arm 3 to determine lineage (appendix p 5). RNA sequencing from whole blood was obtained for 23 patients from study arm 3, who were also part of the COMBAT study,13 a multiomic blood study of patients with COVID-19 (South Central Oxford C Research Ethics Committee in England: Sepsis Immunomics REC reference 19/SC/0296; ISARIC WHO Clinical Characterisation Protocol for Severe Emerging Infections Research Ethics Committee reference 13/SC/0149). Written informed consent was obtained from adults or their personal or nominated consultees for patients lacking capacity, with retrospective consent obtained from the patient once capacity was regained.
Study arm 4 (further external validation of the prognostic value of C19-RS in different populations) comprised 134 patients from the University Hospitals of Leicester NHS Trust and Royal United Hospitals Bath NHS Trust who were verified as SARS-CoV-2 positive and all had a CTPA during their hospital stay, of whom 104 were included in the external validation analysis (appendix p 18). In-hospital short-term outcomes were collected from local hospital electronic patient records. These patients also belonged to the UK C19-CRC substudy of the ORFAN cohort.
Tissue collection, RNA isolation, and sequencing data analysis
In study arm 1, internal mammary artery specimens were collected during surgery and stored at –80°C until processed. Details of tissue collection, RNA extraction, and RNA sequencing are provided in the appendix (pp 2–3).
Defining inflammatory-related genes
To stratify the cohort of patients in study arm 1 by upregulation of inflammatory pathways within the vasculature, relevant to the cytokine storm described in COVID-19, we compiled a list of genes relevant to inflammation annotated in the Gene Ontology (GO) terms “Cytokine Production” (GO:0001816) from the GO main domain “Biological Process” and “Cytokine Activity” (GO:005125) from the GO main domain “Molecular Function”. This list included a range of genes to best represent those involved in the inflammatory response, including C-C motif chemokine ligands, interferons, interleukins, and tumour necrosis factor (TNF) family genes. The full list of the 145 genes selected is provided in the appendix (p 32).
Clustering and differential expression analysis
The set of inflammatory genes was extracted from the count table, filtered to exclude those genes with low expression levels and used to perform unsupervised hierarchical clustering in the study arm 1 population (appendix pp 3–5). The clusters produced were used as the ground truth in a machine learning exercise to build the CT-radiomic signature C19-RS from PVAT, to non-invasively detect this type of cytokine-induced arterial inflammation (figure 1). The same set of genes was extracted from the COMBAT whole-blood-normalised gene expression for patients with a CT scan in order to correlate any potential systemic transcriptional changes with C19-RS.
Adipose tissue segmentation, radiomic characterisation, and machine learning modelling
The detailed coronary and pulmonary CT angiography acquisition protocols for image acquisition are presented in the appendix (pp 5–7). Perivascular adipose tissue segmentation was done manually around the right internal mammary artery from the level of the aortic arch to 120 mm caudally and around the descending thoracic aorta from the level of the pulmonary artery bifurcation to 67·5 mm caudally, as previously described.14 To limit our analysis to radiomic features that could be of value as robust imaging biomarkers, we did rigorous filtering (figure 1), which left a selection of 33 final features used to train the C19-RS signature (appendix p 10). The selection process as well as the full details of the extreme gradient boosting algorithm used to produce C19-RS are presented in the appendix (pp 10–11).
Statistical analysis
Details of descriptive and between-group comparison statistics, power calculations, and handling of missing data with imputation are given in the appendix (pp 11–12). Methods for inter-observer agreement and radiomics quality scoring are available in the appendix (pp 9, 12). A detailed description of all statistical analyses used in each study arm is also presented in the appendix (pp 12–14). When C19-RS was measured, the outcomes data were collected and the statistical analysis took place as a post-hoc investigation of prospectively collected data. Model development and reporting followed TRIPOD (transparent reporting of a multivariable prediction model for individual prediction or diagnosis) guidelines.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or the decision to submit the manuscript for publication.
Results
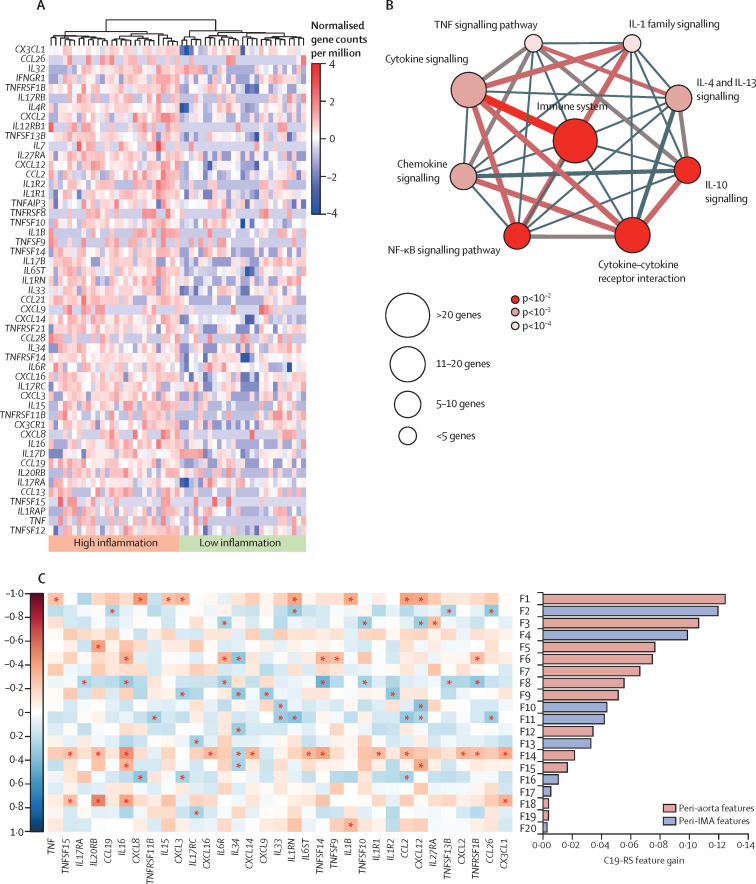
Using unsupervised hierarchical clustering, we identified two clusters of patients with high and low inflammatory cytokine activation within the arterial wall (figure 2A ). Key demographic and clinical characteristics for the two clusters are presented in the appendix (p 33). Pathway enrichment analysis with the top differentially upregulated genes between the two clusters revealed pathways directly related to the immune system and cytokine signalling (figure 2B).
Figure 2.
Unsupervised hierarchical clustering of cytokine production genes expressed in human internal mammary arteries in the study arm 1 population
(A) Unsupervised hierarchical clustering of the list of genes relevant to inflammation annotated in the Gene Ontology (GO) terms “Cytokine Production” (GO:0001816) from the GO main domain “Biological Process” and “Cytokine Activity” (GO:005125) from the GO main domain “Molecular Function”. Hierarchical clustering was done with Ward's method and Minkowski distance, with the Minkowski distance metric, p, set to 10. (B) Enriched signalling pathways of differentially expressed genes between the two clusters of vascular inflammation identified through ConsensusPathDB. (C) Feature importance of the top 20 radiomic features comprising C19-RS and their correlation with cpm (count per million) values with key inflammatory genes in the study arm 1 population (the asterisk denotes significance, p<0·05 by the Spearman's ρ correlation coefficient). An index of the radiomic features included in the figure is presented in the appendix (p 41). The full list of the 145 genes selected is also provided in the appendix (p 32). IMA=internal mammary artery.
Following multiple filtering steps (described in the appendix p 10 and figure 1), a final set of 33 radiomic features was used to train an XGBoost (version 1.4.1.1) machine learning algorithm to quantify cytokine-related arterial inflammation. The product of the algorithm was named C19-RS and retained 25 features (appendix pp 19, 41). To understand the biological interpretation of C19-RS, we tested the relation of each chosen radiomic feature with key inflammatory genes in the arterial wall. Most C19-RS features were differentially associated with multiple genes relevant to cytokine production within the arterial wall (figure 2C). C19-RS had excellent inter-observer agreement among two independent operators (appendix p 9) and achieved a radiomics quality score (RQS) of 77·8%,15 as outlined in the appendix (pp 12, 34–35).
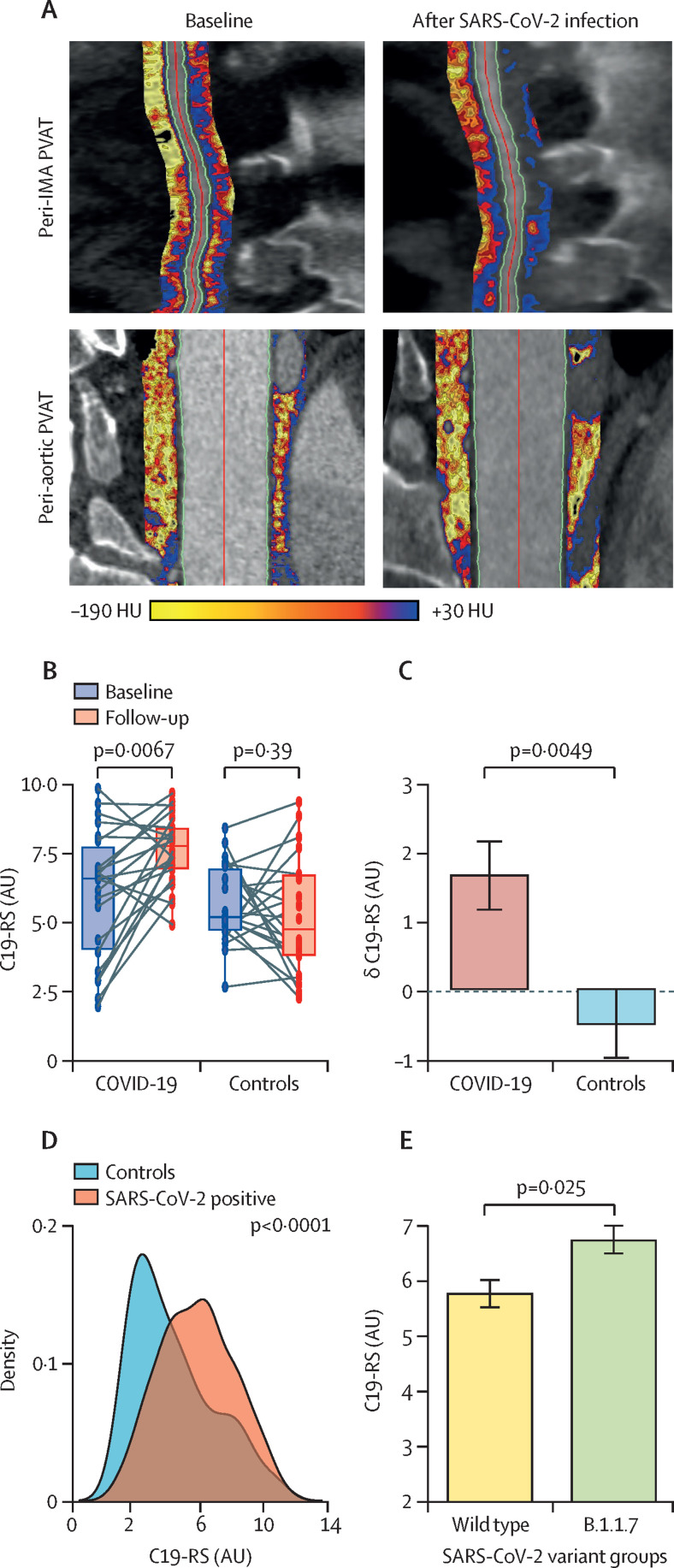
22 patients who had a CCTA before the COVID-19 pandemic (ie, before March, 2020) underwent repeat CCTA less than 6 months after SARS-CoV-2 infection. 22 controls (matched for age, sex, and BMI) who had paired CCTA scans before the COVID-19 pandemic were also identified, and C19-RS was determined in all 88 paired scans (figure 3A ). Demographics are provided in the appendix (p 36). We observed that C19-RS was increased in the COVID-19 group but not in the control group (figure 3B). Indeed, the δ(C19-RS) was strikingly different in patients with COVID-19 versus control patients (p=0·0049; figure 3C).
Figure 3.
C19-RS for COVID-19 detection
(A) Illustration of PVAT mapping in CCTA in a patient 3 years before and during SARS-CoV-2 infection. (B) C19-RS was significantly higher in patients who developed COVID-19 compared to baseline scans, whereas in matched paired controls C19-RS showed no significant change over time. Data are presented as box plots (medians and IQRs). (C) Comparison of the δ values in C19-RS between baseline and follow-up. Data are presented as means (SEs). (D) C19-RS values were higher in SARS-CoV-2-positive patients, with an area under the curve for COVID-19 detection of 0·66 (95% CI 0·59–0·74, p<0·001). (E) Patients with the B.1.1.7 SARS-CoV-2 variant after viral genome sequencing had significantly higher C19-RS values than those infected with the wild-type SARS-CoV-2 variant, suggesting higher degrees of vascular inflammation (data presented as means [SEs] for visualisation). Comparisons made by Wilcoxon signed-rank test in panel B and by Mann-Whitney U test in all other panels. AU=abstract units. CCTA=coronary CT angiogram. HU=Hounsfield units. IMA=internal mammary artery. PVAT=perivascular adipose tissue.
To further test whether C19-RS is applicable in different types of CT angiograms, we applied the signature in non-gated pulmonary artery CT angiograms often done during the acute phase of SARS-CoV-2 infection, in study arm 3 (appendix p 37). C19-RS was significantly higher in SARS-CoV-2-positive patients (on the basis of CT angiograms performed during their hospital admission) compared to SARS-CoV-2-negative patients admitted to hospital during the same period (figure 3D). An optimal cutoff point for the detection of SARS-CoV-2-positive patients was determined by Youden's index (C19-RS 4·21, appendix p 20); patients with C19-RS above that cutoff (n=237) had an adjusted odds ratio (OR) of 2·97 (95% CI 1·43–6·27, p=0·0038) of being SARS-CoV-2 positive compared to patients with low C19-RS values (n=94), after adjusting for age older than 65 years, sex, cardiovascular risk factors (hypertension, hyperlipidaemia, diabetes, BMI, and the presence of coronary artery disease), plasma C-reactive protein (CRP) plasma concentrations, white blood cell count, plasma troponin concentrations, history of chronic obstructive pulmonary disease, and CT scan tube voltage. Addition of the top third (n=8) of radiomic features comprising C19-RS significantly improved the discrimination of a baseline model consisting of the aforementioned covariates for COVID-19 diagnosis (p=0·0066; appendix p 21).
To study the ability of different SARS-CoV-2 variants to cause different degrees of vascular inflammation in line with the severity of disease, we used viral whole-genome sequencing to determine infecting lineages in study arm 3. 115 patients underwent viral whole-genome sequencing, with 71 patients found to have the B.1.1.7 (alpha) variant. Indeed, C19-RS had significantly higher values in patients with the B.1.1.7 variant than in those who were infected with the wild-type SARS-CoV-2 variant (p=0·025; figure 3E). C19-RS was able to distinguish patients infected with the B.1.1.7 variant from those infected with non-B.1.1.7 variants even after adjusting for age, sex, the presence of coronary artery disease, history of chronic obstructive pulmonary disease, white blood cell count, plasma CRP plasma concentrations, and plasma troponin concentrations (adjusted OR 1·89 [95% CI 1·17–3·20] per SD, p=0·012).
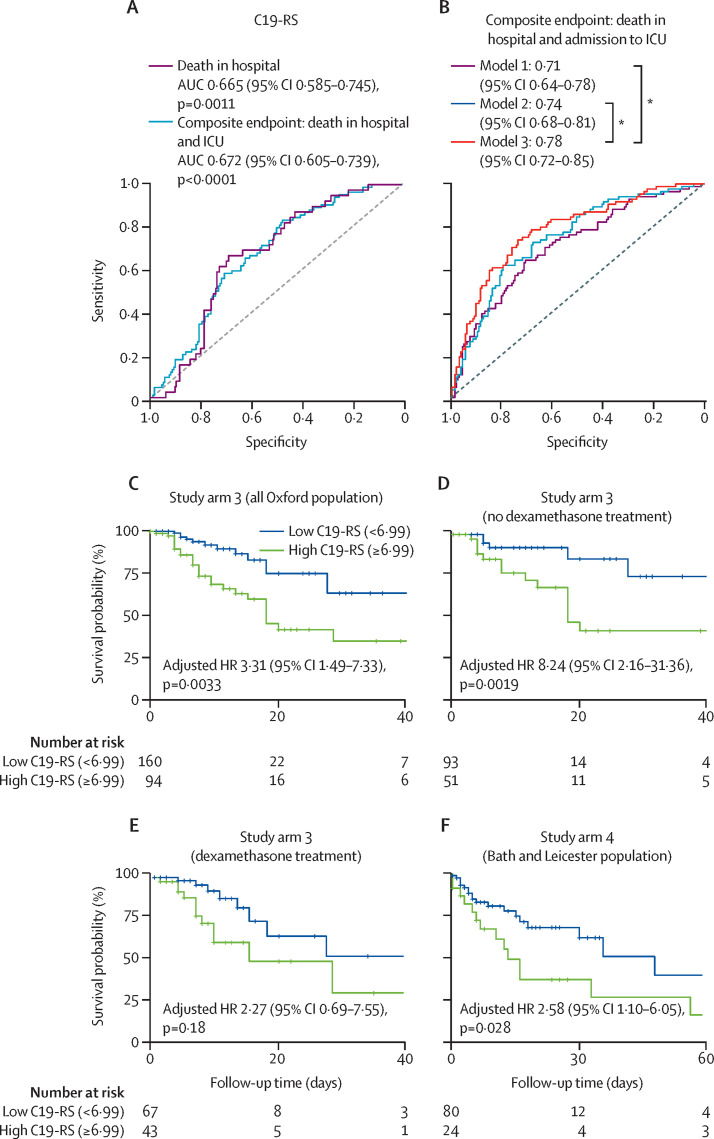
39 SARS-CoV-2-positive patients in study arm 3 died in hospital, and 84 had a composite endpoint of either death in hospital or admission to the intensive care unit (appendix p 18). C19-RS was able to predict both death in hospital (area under the curve [AUC] 66·5%) and the composite endpoint (AUC 67·2%), even when it was used as a single variable (figure 4A ). C19-RS could predict death in hospital with a hazard ratio of 1·91 (95% CI 1·23–2·96, p=0·0036) per SD increase, adjusted for clinical and biochemical risk factors and tube voltage. Based on an optimal cutoff point (C19-RS 6·99, appendix p 22), patients with high C19-RS values (≥6·99) had a 3·3-times higher risk of in-hospital death than patients with low C19-RS values (<6·99; figure 4C). The addition of C19-RS to a comprehensive baseline model consisting of clinical and biochemical risk factors as well as tube voltage significantly improved the accuracy of the model (δAUC=0·04, pDeLong=0·046, panova<0·0001) for predicting the composite endpoint in SARS-CoV-2-positive patients (figure 4B). A decision curve analysis for the composite endpoint is shown in the appendix (p 23). As expected, C19-RS was significantly correlated with plasma CRP concentrations (ρ=0·18, p=0·0033), but the weak correlation coefficient confirms that vascular inflammation is poorly captured by measuring systemic inflammatory biomarkers. C19-RS was also positively related to the length of hospital stay (ρ=0·20, p=0·0017). In a subgroup analysis, we identified patients with no apparent myocardial damage (high sensitivity troponin <20 ng/L), and patients with plasma CRP less than 50 mg/L. Details of missing data are provided in the appendix (p 43). As shown in the appendix (p 42), among 182 patients with no apparent myocardial injury, a high rate of death and the composite endpoint was seen, with 61 (33·5%) of 182 patients identified to have a high degree of vascular inflammation as measured with C19-RS. Among patients with low troponin, there were 16 deaths, 11 of which could have been predicted by C19-RS when used as a single variable, while among those with low CRP (<50mg/L) there were nine deaths, five of which happened among patients with C19-RS above the 6·99 cutoff that defines patients as high risk for in-hospital mortality. Therefore, it appears that even in patients who do not acutely have any cardiovascular complications and have no increase in plasma inflammatory biomarkers, vascular inflammation might still be present and affect clinical outcomes.
Figure 4.
Prognostic value of C19-RS
(A) Univariate receiver operating characteristic (ROC) analysis for the ability of C19-RS to predict death in hospital and a composite endpoint of death in-hospital or intensive care unit (ICU) admission, or both, in the SARS-CoV-2-positive study arm 3 population (n=254). (B) Comparison of ROCs derived from logistic regression models showcasing the additive value of C19-RS in the SARS-CoV-2-positive study arm 3 population (n=254). Model 1 consists of demographic variables (age, sex, hypertension, hyperlipidaemia, diabetes, BMI, presence of coronary artery disease, and history of chronic obstructive pulmonary disease), and tube voltage. Model 2 includes, in addition to the variables in model 1, biochemistry biomarkers (white blood cell count, C-reactive protein, and plasma troponin). Model 3 includes all parameters in model 2 plus C19-RS. (C) Kaplan–Meier curve and adjusted hazard ratio (HR) for in-hospital death for high versus low C19-RS groups in the SARS-CoV-2-positive study arm 3 population (n=254; n=139 from the first wave and n=115 from the second wave) with 39 deaths. HR adjusted for age older than 65 years, sex, cardiovascular risk factors (hypertension, hyperlipidaemia, diabetes, BMI, and presence of coronary artery disease), C-reactive protein plasma concentrations, white blood cell count, plasma troponin, history of chronic obstructive pulmonary disease, tube voltage, and dexamethasone treatment. (D) Kaplan–Meier curve and adjusted HR for in-hospital death for high versus low C19-RS groups in the SARS-CoV-2-positive study arm 3 population that did not receive dexamethasone treatment (n=144 with 19 deaths). (E) Kaplan–Meier curve and adjusted HR for in-hospital death for high versus low C19-RS groups in the SARS-CoV-2-positive study arm 3 population that received dexamethasone treatment (n=110 with 20 deaths). HRs in panels D and E adjusted for age older than 65 years, sex, cardiovascular risk factors (hypertension, hyperlipidaemia, diabetes, and BMI), C-reactive protein plasma concentrations, white blood cell count, history of chronic obstructive pulmonary disease, and tube voltage. (F) Kaplan–Meier curve and adjusted HR for in-hospital death for high versus low C19-RS groups in the external validation study arm 4 population (n=104 with 34 deaths). HR adjusted for age older than 65 years, sex, cardiovascular risk factors (hypertension, hyperlipidaemia, diabetes, BMI, and presence of coronary artery disease), C-reactive protein plasma concentrations, white blood cell count, plasma troponin, history of chronic obstructive pulmonary disease, and tube voltage. *pDeLong value less than 0·05 for AUC comparisons.
The inclusion of patients from the second wave of the pandemic in the UK in study arm 3 (October, 2020, to January, 2021) provided an opportunity to do subgroup analyses based on treatment strategies deployed while the pandemic was still unfolding. Dexamethasone was not part of standard treatment during the first months of the pandemic; however, it became an essential element of COVID-19 management in the second wave. We therefore did a sensitivity analysis with patients receiving dexamethasone (appendix p 14). In the sensitivity analysis with patients receiving dexamethasone, we observed that the prognostic value of C19-RS was decreased (adjusted hazard ratio [HR] 2·27 [p=0·18] vs 8·24 [p=0·0019] in patients receiving dexamethasone vs those not receiving it; figure 4D, E).
Since generalisability is a limitation of machine learning models, we next evaluated the robustness of C19-RS as a prognostic imaging biomarker of arterial inflammation in COVID-19 in a third independent study arm (study arm 4), comprising 104 consecutive SARS-CoV-2-positive patients from different locations with different local demographics from Bath and Leicester (appendix p 38). 34 in-hospital deaths were recorded, while the composite endpoint was reached in 60 patients (appendix p 18). C19-RS was associated with mortality, even after adjusting for clinical and biochemical risk factors and tube voltage (adjusted HR 1·61 [95% CI 1·08–2·40] per SD increase, p=0·019). Using the optimal cutoff point calculated in study arm 3 (C19-RS 6·99), patients with high C19-RS values had a 2·6-times higher risk of in-hospital death than patients with low C19-RS values after aforementioned adjustments (figure 4F). Patients with high C19-RS values also had a 4·9-times higher risk of meeting the composite endpoint than those with low C19-RS values (adjusted OR 4·92 [95% CI 1·54–18·02, p=0·0104). This provides an external validation of C19-RS with different scanners and hospital settings in patients from different geographical locations than those in study arm 3 (appendix pp 5–7).
Training of C19-RS included gated coronary CTAs and validation included non-gated pulmonary CTAs, involving two different imaging phases and scan settings. However, C19-RS analysis is done in the perivascular space, and it does not correlate with the lumen attenuation (appendix p 30). Addition of rater as a covariate in our multivariable models had no impact on the significance or effect size of C19-RS on the prediction of in-hospital outcomes (the adjusted HR for high C19-RS in predicting in-hospital mortality was 3·20 [95% CI 1·42–7·19], p=0·0049 in study arm 3 and 2·85 [1·20–6·75], p=0·013 in study arm 4; appendix p 9). Additionally, there were no significant differences in C19-RS values between CCTA and CTPAs (p=0·21; appendix p 31). Sensitivity analyses for our prognostic models excluding variables with missingness above 10% showed similar results (appendix p 43).
To further explore the cause of vascular inflammation detected by C19-RS, we examined the gene expression profiles of whole blood from 23 patients. We first examined the expression of the inflammatory gene set (77 inflammatory genes expressed in whole blood). No relationship was found between the expression profile of these genes and the C19-RS score (appendix pp 25–26), nor were any genes significantly differentially expressed (false discovery rate-adjusted p<0·05) between patients with a high or low C19-RS score in a global analysis, suggesting that C19-RS is a vessel-centric inflammatory signature, not captured by measurements of systemic inflammation (appendix p 24). However, when analysing the correlation of C19-RS with eigengenes summarising gene coexpression modules derived with weighted gene correlation network analysis on the full COMBAT dataset (appendix pp 25–26), we found a correlation between C19-RS and the MEgreen eigengene module (r=0·61, p=0·00031; appendix pp 25–26, 44–64), which is highly enriched in coagulation pathways (appendix pp 25–26, 65–67).
Discussion
We present a new platform that uses quantitative radiotranscriptomics to measure cytokine-induced vascular inflammation from routine CT angiograms of the coronary or pulmonary arteries. A radiotranscriptomic signature (C19-RS) was developed with radiomic analysis of the perivascular space around the internal mammary arteries and the aorta, and it was trained against the RNA-sequencing transcriptomic fingerprint of vascular cytokine-driven inflammation of internal mammary artery biopsies. C19-RS was found to significantly increase after SARS-CoV-2 infection, and its increase was related to high activation of prothrombotic pathways in the circulating blood, and increased risk of in-hospital mortality. Treatment with dexamethasone seems to be effective in reducing the vascular inflammatory risk of in-hospital mortality in patients with COVID-19. These findings suggest that radiotranscriptomic biomarkers could be used to inform deployment of anti-inflammatory treatments to those patients who need them, in COVID-19 and beyond. The links between C19-RS and coagulation (but not with inflammatory signatures in peripheral blood) imply that vascular inflammation might be the trigger of thrombosis, which is responsible for multiorgan failure in COVID-19. This provides an important mechanistic insight into COVID-19 pathogenesis.
Acute cytokine-driven inflammation observed in COVID-19 leads to microthrombosis in pulmonary as well as systemic circulation, contributing to hypoxic respiratory failure from acute respiratory distress syndrome16 and multiorgan failure.17, 18 SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2) receptor, expressed in the vasculature,19 triggering a cytokine storm in the arterial wall3 that increases vascular permeability and changes the texture of perivascular structures, including PVAT. Quantifying cytokine-induced vascular inflammation in patients with COVID-19 is an unmet need not only for risk stratification of patients admitted to hospital to optimise management and allocation of resources, but also for informing deployment of preventive measures after hospital discharge, as viral infections have been known to increase the risk of future cardiovascular events.20
Vessel-derived cytokines diffuse into the perivascular space and cause characteristic changes in the texture of the perivascular space, which can be captured and quantified in clinical CT angiography using texture radiomics.21 By using a radiotranscriptomic approach, we constructed a novel imaging signature, the C19-RS, which can be extracted from any CT angiogram that visualises the internal mammary arteries, and provides a quantitative measure of cytokine-induced vascular inflammation in patients with acute SARS-CoV-2 infection. The power of this radiotranscriptomic approach is highlighted by the fact that the C19-RS was trained with gated coronary CT angiograms from patients without COVID-19 and was then tested in CT angiograms of the pulmonary artery, in patients with COVID-19 from three different geographical areas in England, with striking prognostic value. This COVID-19-related inflammatory risk was found to respond well to dexamethasone treatment, suggesting that it could be used to inform deployment of steroids to those who need them. This is important as corticosteroids increase the risk of mucormycosis, a rare fungal infection with a 30·7% mortality rate in patients with COVID-19,22 so imaging-guided prescription of these drugs could have a major impact in the management of these patients. Furthermore, as C19-RS is a generic signature of cytokine-driven vascular inflammation, it is expected to be applicable in other diseases beyond COVID-19. This study also shows that platforms based on quantitative radiotranscriptomics can offer unique opportunities for rapid development of custom-made, non-invasive imaging biomarkers that describe the underlying vascular biology against which they have been trained. Although other studies have used radiomics-based analyses of the lungs to predict COVID-19 outcomes,23, 24, 25, 26 this study is, to the best of our knowledge, the first to apply a radiotranscriptomic approach to quantify vascular inflammation in COVID-19, and paves the way for wider adoption of this approach in the search for novel imaging biomarkers. Future directions could involve application of this approach in different types of vascular inflammatory diseases, such as aortic aneurysms, carotid artery disease, or even autoimmune diseases such as temporal arteritis. This approach also offers unique opportunities for the management of future outbreaks of novel SARS-CoV-2 variants or other infectious disease outbreaks.
The limitations of this study include the fact that the effect of dexamethasone on the vascular inflammatory risk defined by C19-RS was not investigated in a randomised clinical trial setting and should therefore be considered as hypothesis generating. Additionally, no race or ethnicity data were included in the adjustments, and viral sequencing was only done for the SARS-CoV-2 B.1.1.7 variant. Moreover, the testing populations in our study were highly selected patients admitted to hospital with acute COVID-19 undergoing CTPA imaging, and the results cannot be extrapolated to the general population with COVID-19. Training and validation of C19-RS included coronary and pulmonary CT angiograms, involving two different imaging phases. Although the different scan settings could have introduced noise in the CTPA analysis, the striking prognostic value of C19-RS extracted from CTPA scans of patients with COVID-19 provides additional evidence for the potential generalisability of the signature. The subanalysis for correlation between C19-RS and the gene expression profiles of whole blood was performed in a small number of patients (n=23), and the sampling timepoint for bulk RNA sequencing is not absolutely contemporaneous, ranging between 12 days before and 6 days after the C19-RS measurement (appendix p 25). Additionally, a subset of patients in study arm 3 was used in one of the radiomic features filtering steps, with the remainder of the study arm 3 population serving as an internal test cohort. Given the real-world setting of this study, we applied imputation for missing values to include datasets that were as complete as possible.
In conclusion, in this study we presented a novel radiotranscriptomic platform that allows development of customised imaging biomarkers of vascular inflammation, tailored to the type of vascular inflammation of interest, and the changes that it causes to the perivascular space. This new platform was tested in patients with COVID-19 by constructing and validating a signature of cytokine-driven vascular inflammation (C19-RS). This signature has prognostic value for COVID-19 outcomes, particularly for in-hospital mortality, and it identifies potential responders to corticosteroids. This technology could be used, pending further validation, to stratify patients after they develop SARS-CoV-2 infection, and it could also potentially be applicable to various vascular inflammatory diseases, where cytokine-driven inflammation is biologically relevant, following appropriate independent validation and cost-effectiveness analyses.
Data sharing
Individual participant-level data used for this report are not publicly available, because they contain protected patient health information. The authors agree to apply the machine learning algorithm to data provided by other academic researchers on their behalf for research purposes only, following completion of a data sharing agreement. Requests for data access should be directed to the corresponding author via email, and will be reviewed by the Publications and Data Sharing Committee of the ORFAN study. Sequence data generated during the study are available from the European Nucleotide Archive under study accession number PRJEB43319.
Declaration of Interests
CA, KC, CS, and SN are founders, shareholders, and directors of Caristo Diagnostics, a CT image analysis company. CS is a full-time employee and MS is a part-time employee of Caristo diagnostics. JD is shareholder and chair of the advisory board of Caristo Diagnostics. EKO is a consultant and minor shareholder of Caristo Diagnostics. The technology described in this work is subject to patent US10,695,023B2 and patent applications PCT/GB2017/053262, GB2018/1818049.7, GR20180100490, and GR20180100510, licensed through exclusive license to Caristo Diagnostics. Caristo Diagnostics and the authors linked to it have no further conflicts of interest, beyond the above. JD is CMO of Our Future Health; Senior Advisor for Cardiovascular Disease Prevention, NHS Healthcheck Expert Scientific and Clinical Advisory Panel; and Chair of the Review of the National Health Check Programme for Public Health England. JCLR received a Research for Patient Benefit Grant from NIHR, and consulting fees from HeartFlow for physician services. DAd received support from Leicester NIHR Biomedical Research Unit and Innovate UK; grants and contracts from the Medical Research Council; and has two patents issued (Cardiac assist device: EP3277337A1; and angioplasty of calcified arteries: PCT/GB2017/050877) outside the scope of the current study. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
Caristo Diagnostics is a spinout company from the University of Oxford, Oxford, UK, that participates in the ORFAN study and its UK COVID-19 Cardiovascular Research Consortium substudy. The company offers its cloud gateway (which allows anonymisation and transfer of the CT scans from the sites to the core laboratory in Oxford) and analysis platforms (CaRi platform) to the ORFAN study, as part of an IP licence agreement. Caristo Diagnostics is also the lead applicant in the Innovate UK grant that supported the building of the analysis platforms (CaRi-Research 2.1.1. toolbox), which are quoted in the appendix (pp 8, 9). Employees of Caristo Diagnostics also offered high-end expertise in deep learning modelling, as part of their formal participation for the British Heart Foundation grants TG/19/2/34831 and RG/F/21/110040. We are grateful to Hannah Cooper, Robert Shaw, and Monique Andersson who helped with acquisition of ventilation data, which were used for scoring COVID-19 severity. We are also grateful to Ioannis Akoumianakis for his contribution in RNA extraction. Lastly, we are grateful to the COMBAT Consortium Members and the ORFAN Investigators. CPK acknowledges support from the EPSRC, the Scatcherd Fund at the University of Oxford and the Onassis Foundation. CA acknowledges support from the British Heart Foundation (CH/F/21/90009, TG/19/2/34831 and RG/F/21/110040), the Oxford BHF Centre of Research Excellence (RE/18/3/34214), the Oxford NIHR Biomedical Research Centre and Innovate UK. KC acknowledges support from the British Heart Foundation (FS/16/15/32047, TG/16/3/32687, CH/16/1/32013), the Oxford BHF Centre of Research Excellence (RE/18/3/34214) and Innovate UK. LPH is supported in part by the Oxford NIHR Biomedical Research Centre. The Wellcome Sanger Institute is supported by core funding from the Wellcome Trust (grant number 206194). The research was also supported by the Wellcome Trust Core Award 203141/Z/16/Z. This work is part of the UK COVID-19 Cardiovascular Research Consortium, a UK National Flagship Programme supported by the British Heart Foundation–National Institute of Health Research (NIHR) Cardiovascular Partnership. SL is a Wellcome Trust Clinical Research Fellow. Finally, the investigators acknowledge the philanthropic support of the donors to the University of Oxford's COVID-19 Research Response Fund.
Acknowledgments
COMBAT Consortium members
David J Ahern, Zhichao Ai, Mark Ainsworth, Chris Allan, Alice Allcock, Brian Angus, M Azim Ansari, Carolina V Arancibia-Cárcamo, Dominik Aschenbrenner, Moustafa Attar, J Kenneth Baillie, Eleanor Barnes, Rachael Bashford-Rogers, Archana Bashyal, Sally Beer, Georgina Berridge, Amy Beveridge, Sagida Bibi, Tihana Bicanic, Luke Blackwell, Paul Bowness, Andrew Brent, Andrew Brown, John Broxholme, David Buck, Katie L Burnham, Helen Byrne, Susana Camara, Ivan Candido Ferreira, Philip Charles, Wentao Chen, Yi-Ling Chen, Amanda Chong, Elizabeth A Clutterbuck, Mark Coles, Christopher P Conlon, Richard Cornall, Adam P Cribbs, Fabiola Curion, Emma E Davenport, Neil Davidson, Simon Davis, Calliope A Dendrou, Julie Dequaire, Lea Dib, James Docker, Christina Dold, Tao Dong, Damien Downes, Hal Drakesmith, Susanna J Dunachie, David A Duncan, Chris Eijsbouts, Robert Esnouf, Alexis Espinosa, Rachel Etherington, Benjamin Fairfax, Rory Fairhead, Hai Fang, Shayan Fassih, Sally Felle, Maria Fernandez Mendoza, Ricardo Ferreira, Roman Fischer, Thomas Foord, Aden Forrow, John Frater, Anastasia Fries, Veronica Gallardo Sanchez, Lucy C Garner, Clementine Geeves, Dominique Georgiou, Leila Godfrey, Tanya Golubchik, Maria Gomez Vazquez, Angie Green, Hong Harper, Heather A Harrington, Raphael Heilig, Svenja Hester, Jennifer Hill, Charles Hinds, Clare Hird, Ling-Pei Ho, Renee Hoekzema, Benjamin Hollis, Jim Hughes, Paula Hutton, Matthew A Jackson-Wood, Ashwin Jainarayanan, Anna James-Bott, Kathrin Jansen, Katie Jeffery, Elizabeth Jones, Luke Jostins, Georgina Kerr, David Kim, Paul Klenerman, Julian C Knight, Vinod Kumar, Piyush Kumar Sharma, Prathiba Kurupati, Andrew Kwok, Angela Lee, Aline Linder, Teresa Lockett, Lorne Lonie, Maria Lopopolo, Martyna Lukoseviciute, Jian Luo, Spyridoula Marinou, Brian Marsden, Jose Martinez, Philippa C Matthews, Michalina Mazurczyk, Simon McGowan, Stuart McKechnie, Adam Mead, Alexander J Mentzer, Yuxin Mi, Claudia Monaco, Ruddy Montadon, Giorgio Napolitani, Isar Nassiri, Alex Novak, Darragh P O’Brien, Daniel O’Connor, Denise O’Donnell, Graham Ogg, Lauren Overend, Inhye Park, Ian Pavord, Yanchun Peng, Frank Penkava, Mariana Pereira Pinho, Elena Perez, Andrew J Pollard, Fiona Powrie, Bethan Psaila, T Phuong Quan, Emmanouela Repapi, Santiago Revale, Laura Silva-Reyes, Jean-Baptiste Richard, Charlotte Rich-Griffin, Thomas Ritter, Christine S Rollier, Matthew Rowland, Fabian Ruehle, Mariolina Salio, Stephen Nicholas Sansom, Raphael Sanches Peres, Alberto Santos Delgado, Tatjana Sauka-Spengler, Ron Schwessinger, Giuseppe Scozzafava, Gavin Screaton, Anna Seigal, Malcolm G Semple, Martin Sergeant, Christina Simoglou Karali, David Sims, Donal Skelly, Hubert Slawinski, Alberto Sobrinodiaz, Nikolaos Sousos, Lizzie Stafford, Lisa Stockdale, Marie Strickland, Otto Sumray, Bo Sun, Chelsea Taylor, Stephen Taylor, Adan Taylor, Supat Thongjuea, Hannah Thraves, John A Todd, Adriana Tomic, Orion Tong, Amy Trebes, Dominik Trzupek, Felicia Anna Tucci, Lance Turtle, Irina Udalova, Holm Uhlig, Erinke van Grinsven, Iolanda Vendrell, Marije Verheul, Alexandru Voda, Guanlin Wang, Lihui Wang, Dapeng Wang, Peter Watkinson, Robert Watson, Michael Weinberger, Justin Whalley, Lorna Witty, Katherine Wray, Luzheng Xue, Hing Yuen Yeung, Zixi Yin, Rebecca K Young, Jonathan Youngs, Ping Zhang, Yasemin-Xiomara Zurke.
ORFAN investigators
Adrian P Banning, Alexios S Antonopoulos, Amrita Bajaj, Andrew David Kelion, Aparna Deshpande, Attila Kardos, Benjamin Hudson, Bon-Kwon Koo, Cheerag Shirodaria, Cheng Xie, Christos Kotanidis, Ciara Mahon, Colin Berry, David Adlam, David Ernest Newby, Derek Leslie Connolly, Diane Elizabeth Scaletta, Donna Alexander, Ed Nicol, Elisa McAlindon, Evangelos Oikonomou, Francesca Pugliese, Gianluca Pontone, Giulia Benedetti, Guo-Wei He, Henry West, Hidekazu Kondo, Imre S Benedek, Intrajeet Das, John Deanfield, John Graby, John Greenwood, Jonathan Carl Luis Rodrigues, Junbo Ge, Keith M Channon, Larissa Fabritz, Li-Juan Fan, Lucy Kingham, Marco Guglielmo, Maria Lyasheva, Matthias Schmitt, Meinrad Beer, Michelle Louise Anderson, Milind Y Desai, Mohamed Marwan, Naohiko Takahashi, Nehal N Mehta, Neng Dai, Nicholas Screaton, Nikant Kumar Sabharwal, Pál Maurovich-Horvat, Praveen PG Rao, Rafail Angelos Kotronias, Rajesh Kumar Kharbanda, Rebecca Louise Preston, Richard James Wood, Ron Blankstein, Ronak Rajani, Saeed Mirsadraee, Shahzad Mahmood Munir, Sheena Thomas, Stefan Neubauer, Steffen Christoph Klömpken, Steffen E Petersen, Stephan Achenbach, Susan Anthony, Sze Mun Mak, Tarun Mittal, Theodora M Benedek, Vinoda Sharma, Wen-Hua Lin.
Contributors
CPK, CX, ST, and EKO were responsible for image analysis. DAl, JCLR, RK, ML, SFL, BC, NS, GR, KKC, AL, MT, and FG were responsible for data collection. ST, VS, SF, GK, and RS were responsible for recruitment of patients and provision of clinical samples. CA, CPK, and IB conceived and performed experiments. JCLR, ST, DAl, PR, DI, AD, AB, and BJH were responsible for image acquisition. CPK, MS, HL, KB, AM, and DO’C were responsible for data analysis. JCLR, BJH, CS, DAd, L-PH, SN, KMC, DEN, JD, JK, and CA were responsible for supervision. CA was responsible for overall project design. CPK, HL, KB, LPH, and CA interpreted data and wrote the manuscript. JCLR, L-PH, DAd, KMC, SN, EKO, CS, DEN, and JD were responsible for critical revision of the manuscript. All authors had access to all the raw data sets. CPK, CX, HL, MS, L-PH, and CA directly accessed and verified the underlying data. CA was responsible for the decision to submit the manuscript for publication.
Contributor Information
COMBAT Consortium:
David J Ahern, Zhichao Ai, Mark Ainsworth, Chris Allan, Alice Allcock, Brian Angus, M Azim Ansari, Carolina Arancibia-Cárcamo, Dominik Aschenbrenner, Moustafa Attar, J Kenneth Baillie, Eleanor Barnes, Rachael Bashford-Rogers, Archana Bashyal, Sally Beer, Georgina Berridge, Amy Beveridge, Sagida Bibi, Tihana Bicanic, Luke Blackwell, Paul Bowness, Andrew Brent, Andrew Brown, John Broxholme, David Buck, Katie Burnham, Helen Byrne, Susana Camara, Ivan Candido Ferreira, Philip Charles, Wentao Chen, Yi-Ling Chen, Amanda Chong, Elizabeth Clutterbuck, Mark Coles, Christopher Conlon, Richard Cornall, Adam Cribbs, Fabiola Curion, Emma Davenport, Neil Davidson, Simon Davis, Calliope Dendrou, Julie Dequaire, Lea Dib, James Docker, Christina Dold, Tao Dong, Damien Downes, Hal Drakesmith, Susanna Dunachie, David Duncan, Chris Eijsbouts, Robert Esnouf, Alexis Espinosa, Rachel Etherington, Benjamin Fairfax, Rory Fairhead, Hai Fang, Shayan Fassih, Sally Felle, Maria Fernandez Mendoza, Ricardo Ferreira, Roman Fischer, Thomas Foord, Aden Forrow, John Frater, Anastasia Fries, Veronica Gallardo Sanchez, Lucy Garner, Clementine Geeves, Dominique Georgiou, Leila Godfrey, Tanya Golubchik, Maria Gomez Vazquez, Angie Green, Hong Harper, Heather Harrington, Raphael Heilig, Svenja Hester, Jennifer Hill, Charles Hinds, Clare Hird, Ling-Pei Ho, Renee Hoekzema, Benjamin Hollis, Jim Hughes, Paula Hutton, Matthew Jackson-Wood, Ashwin Jainarayanan, Anna James-Bott, Kathrin Jansen, Katie Jeffery, Elizabeth Jones, Luke Jostins, Georgina Kerr, David Kim, Paul Klenerman, Julian Knight, Vinod Kumar, Piyush Kumar Sharma, Prathiba Kurupati, Andrew Kwok, Angela Lee, Aline Linder, Teresa Lockett, Lorne Lonie, Maria Lopopolo, Martyna Lukoseviciute, Jian Luo, Spyridoula Marinou, Brian Marsden, Jose Martinez, Philippa Matthews, Michalina Mazurczyk, Simon McGowan, Stuart McKechnie, Adam Mead, Alexander Mentzer, Yuxin Mi, Claudia Monaco, Ruddy Montadon, Giorgio Napolitani, Isar Nassiri, Alex Novak, Darragh O'Brien, Daniel O'Connor, Denise O'Donnell, Graham Ogg, Lauren Overend, Inhye Park, Ian Pavord, Yanchun Peng, Frank Penkava, Mariana Pereira Pinho, Elena Perez, Andrew Pollard, Fiona Powrie, Bethan Psaila, T Phuong Quan, Emmanouela Repapi, Santiago Revale, Laura Silva-Reyes, Jean-Baptiste Richard, Charlotte Rich-Griffin, Thomas Ritter, Christine Rollier, Matthew Rowland, Fabian Ruehle, Mariolina Salio, Stephen Nicholas Sansom, Raphael Sanches Peres, Alberto Santos Delgado, Tatjana Sauka-Spengler, Ron Schwessinger, Giuseppe Scozzafava, Gavin Screaton, Anna Seigal, Malcolm Semple, Martin Sergeant, Christina Simoglou Karali, David Sims, Donal Skelly, Hubert Slawinski, Alberto Sobrinodiaz, Nikolaos Sousos, Lizzie Stafford, Lisa Stockdale, Marie Strickland, Otto Sumray, Bo Sun, Chelsea Taylor, Stephen Taylor, Adan Taylor, Supat Thongjuea, Hannah Thraves, John Todd, Adriana Tomic, Orion Tong, Amy Trebes, Dominik Trzupek, Felicia Anna Tucci, Lance Turtle, Irina Udalova, Holm Uhlig, Erinke van Grinsven, Iolanda Vendrell, Marije Verheul, Alexandru Voda, Guanlin Wang, Lihui Wang, Dapeng Wang, Peter Watkinson, Robert Watson, Michael Weinberger, Justin Whalley, Lorna Witty, Katherine Wray, Luzheng Xue, Hing Yuen Yeung, Zixi Yin, Rebecca Young, Jonathan Youngs, Ping Zhang, and Yasemin-Xiomara Zurke
ORFAN investigators:
Adrian Banning, Alexios Antonopoulos, Amrita Bajaj, Andrew Kelion, Aparna Deshpande, Attila Kardos, Benjamin Hudson, Bon-Kwon Koo, Cheerag Shirodaria, Cheng Xie, Christos Kotanidis, Ciara Mahon, Colin Berry, David Adlam, David Newby, Derek Connolly, Diane Scaletta, Donna Alexander, Ed Nicol, Elisa McAlindon, Evangelos Oikonomou, Francesca Pugliese, Gianluca Pontone, Giulia Benedetti, Guo-Wei He, Henry West, Hidekazu Kondo, Imre Benedek, Intrajeet Das, John Deanfield, John Graby, John Greenwood, Jonathan Rodrigues, Junbo Ge, Keith Channon, Larissa Fabritz, Li-Juan Fan, Lucy Kingham, Marco Guglielmo, Maria Lyasheva, Matthias Schmitt, Meinrad Beer, Michelle Anderson, Milind Desai, Mohamed Marwan, Naohiko Takahashi, Nehal Mehta, Neng Dai, Nicholas Screaton, Nikant Sabharwal, Pál Maurovich-Horvat, Praveen Rao, Rafail Kotronias, Rajesh Kharbanda, Rebecca Preston, Richard Wood, Ron Blankstein, Ronak Rajani, Saeed Mirsadraee, Shahzad Munir, Sheena Thomas, Stefan Neubauer, Steffen Klömpken, Steffen Petersen, Stephan Achenbach, Susan Anthony, Sze Mak, Tarun Mittal, Theodora Benedek, Vinoda Sharma, and Wen-Hua Lin
Supplementary Material
References
- 1.Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol. 2020;20:389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterer GW, Rello J. Steroids and COVID-19: we need a precision approach, not one size fits all. Infect Dis Ther. 2020;9:701–705. doi: 10.1007/s40121-020-00338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aghamohammadzadeh R, Withers S, Lynch F, Greenstein A, Malik R, Heagerty A. Perivascular adipose tissue from human systemic and coronary vessels: the emergence of a new pharmacotherapeutic target. Br J Pharmacol. 2012;165:670–682. doi: 10.1111/j.1476-5381.2011.01479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonopoulos AS, Sanna F, Sabharwal N, et al. Detecting human coronary inflammation by imaging perivascular fat. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal2658. [DOI] [PubMed] [Google Scholar]
- 7.Kotanidis CP, Antoniades C. Perivascular fat imaging by computed tomography (CT): a virtual guide. Br J Pharmacol. 2021;178:4270–4290. doi: 10.1111/bph.15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oikonomou EK, Antoniades C. The role of adipose tissue in cardiovascular health and disease. Nat Rev Cardiol. 2019;16:83–99. doi: 10.1038/s41569-018-0097-6. [DOI] [PubMed] [Google Scholar]
- 9.Oikonomou EK, Williams MC, Kotanidis CP, et al. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J. 2019;40:3529–3543. doi: 10.1093/eurheartj/ehz592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotanidis CP, Antoniades C. In: Epicardial adipose tissue: from cell to clinic. Iacobellis G, editor. Springer International Publishing; Cham: 2020. Perivascular adipose tissue and atherosclerosis; pp. 91–115. [Google Scholar]
- 11.Otsuka F, Yahagi K, Sakakura K, Virmani R. Why is the mammary artery so special and what protects it from atherosclerosis? Ann Cardiothorac Surg. 2013;2:519–526. doi: 10.3978/j.issn.2225-319X.2013.07.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akoumianakis I, Sanna F, Margaritis M, et al. Adipose tissue-derived WNT5A regulates vascular redox signaling in obesity via USP17/RAC1-mediated activation of NADPH oxidases. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahern DJ, Ai Z, Ainsworth M, et al. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185:916–938. doi: 10.1016/j.cell.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlett CL, Massaro JM, Lehman SJ, et al. Novel measurements of periaortic adipose tissue in comparison to anthropometric measures of obesity, and abdominal adipose tissue. Int J Obes. 2009;33:226–232. doi: 10.1038/ijo.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 16.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 17.Guzik TJ, Mohiddin SA, Dimarco A, et al. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Du Z, Zhu F, et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395:e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow EJ, Rolfes MA, O'Halloran A, et al. Acute cardiovascular events associated with influenza in hospitalized adults: a cross-sectional study. Ann Intern Med. 2020;173:605–613. doi: 10.7326/M20-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotanidis CP, Antoniades C. Selfies in cardiovascular medicine: welcome to a new era of medical diagnostics. Eur Heart J. 2020;41:4412–4414. doi: 10.1093/eurheartj/ehaa608. [DOI] [PubMed] [Google Scholar]
- 22.Singh AK, Singh R, Joshi SR, Misra A. Mucormycosis in COVID-19: a systematic review of cases reported worldwide and in India. Diabetes Metab Syndr. 2021;15 doi: 10.1016/j.dsx.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q, Wang S, Li L, et al. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10:7231–7244. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Wang D, Shao J, et al. A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography. Sci Rep. 2021;11 doi: 10.1038/s41598-021-83237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiri I, Sorouri M, Geramifar P, et al. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput Biol Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh R, Elsakka A, Wray R, et al. FDG PET/CT imaging features and clinical utility in COVID-19. Clin Imaging. 2021;80:262–267. doi: 10.1016/j.clinimag.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual participant-level data used for this report are not publicly available, because they contain protected patient health information. The authors agree to apply the machine learning algorithm to data provided by other academic researchers on their behalf for research purposes only, following completion of a data sharing agreement. Requests for data access should be directed to the corresponding author via email, and will be reviewed by the Publications and Data Sharing Committee of the ORFAN study. Sequence data generated during the study are available from the European Nucleotide Archive under study accession number PRJEB43319.