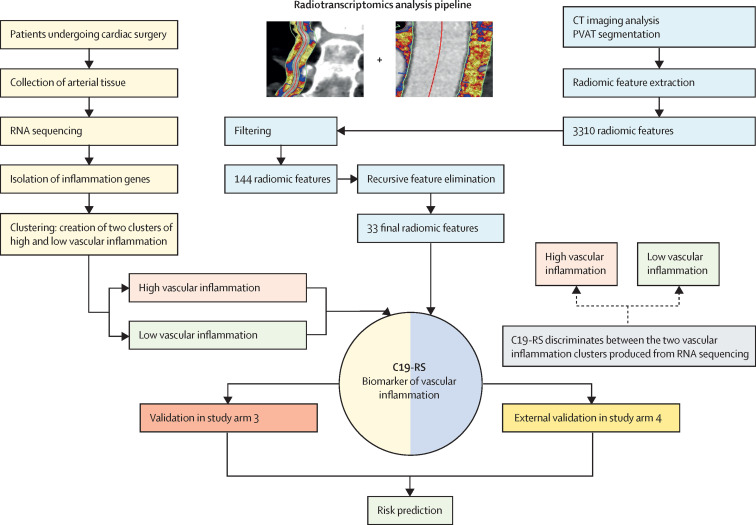
Figure 1.
Workflow for building the radiotranscriptomic signature C19-RS
Workflow depicting the multiple steps taken to develop the radiomic signature C19-RS. To limit our analysis to radiomic features that could be of value as imaging biomarkers, we did a series of filtering steps, to exclude features that are not stable in test–retest analyses, features that are highly correlated with each other, and features that are significantly correlated with BMI or intrathoracic adipose tissue volume, to retain only features that predict the outcome variable with the same direction within study arm 1 and 20% of the exploratory study arm 3 subpopulation. Finally, recursive feature elimination with a random forest algorithm and repeated five-times cross-validation showed a plateau in the accuracy of the trained model with 33 final features. Those features were next used within the study arm 1 population to train an XGBoost algorithm using decisions trees in order to identify patients with activated inflammatory pathways within their arterial vasculature. The raw product of the algorithm was named C19-RS. PVAT=perivascular adipose tissue.