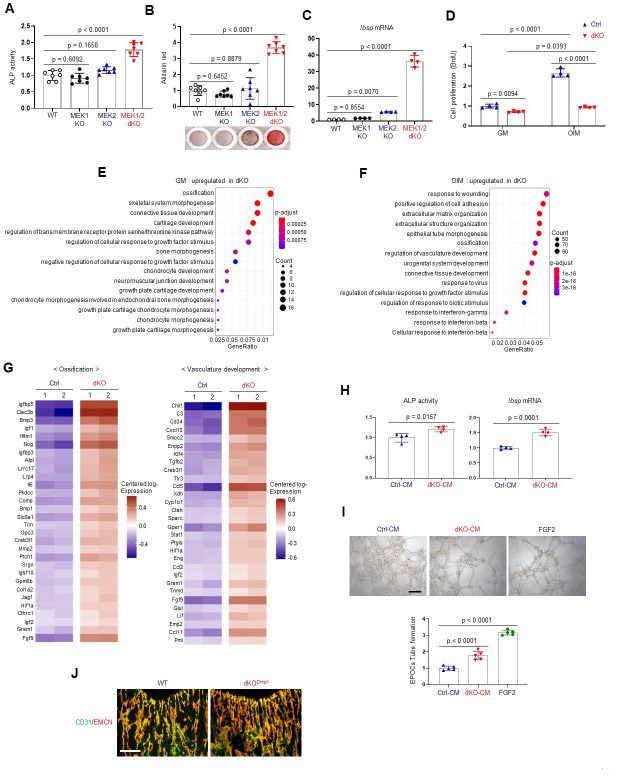
Figure 3. Effects of MEK1/2 deletion on osteoblast differentiation and production of angiogenic and osteogenic factors.
(A–C) Mouse Map2k1fl/fl;Map2k2+/+ and Map2k1fl/fl;Map2k2−/− osteoblasts (Obs) were infected with control vector or Cre recombinase-expressing lentiviruses; Map2k1fl/fl;Map2k2+/+ Obs with control (WT) or Cre (MEK1 KO), Map2k1fl/fl;Map2k2−/− Obs with control (MEK2 KO) or Cre (MEK1/2 dKO). Puromycin-selected Obs were cultured under osteogenic conditions and alkaline phosphatase (ALP) activity (A) and osteogenic gene expression (C) were determined at day 6 and mineralization (B) was analyzed at day 18 of culture. (D) MEK2 KO (Ctrl) and MEK1/2 dKO (dKO) Obs were cultured with control growth medium (GM) or osteogenic induction medium (OIM) and cell proliferation was analyzed by Bromodeoxyuridine (BrdU) incorporation at day 6 of the culture. (E, F) Transcriptome analysis of Ctrl and dKO Obs 6 days after GM or OIM culture. Biological process output of gene ontology analysis was performed in both GM and OIM group for upregulated genes in dKO relative to Ctrl Obs. The color indicates adjusted p value as estimated by the Benjamini–Hochberg method with the threshold of significance p = 0.05 and q = 0.005. (G) Heatmaps for ossification- and vasculature-associated gene expression. The top 30 upregulated genes in dKO Obs relative to Ctrl Obs are displayed as each row and column represent gene symbol and sample, respectively. The log10 expression (read count) was centered across samples and red and purple denote upregulated and downregulated, respectively. (H) Conditioned medium (CM) from Ctrl and dKO Obs was collected at day 6 under osteogenic culture and mouse wildtype bone marrow-derived mesenchymal stromal cells (BMSCs) were cultured under osteogenic condition in the presence of CM of Ctrl Obs and dKO Obs and ALP activity (left) and Ibsp mRNA level (right) were assessed at day 6. (I) Capillary tube formation of mouse endothelial cells (EPOCs) was performed in the presence of CM for 5 hr. Representative images (top) and quantification for the number of branches are displayed (bottom). FGF2 was used as a positive control. Scale bar, 200 µm. (J) Immunofluorescence for CD31 (green) and endomucin (EMCN, red) in the epiphyseal area of 8-week-old WT and dKODmp1 femurs. Scale bar, 100 µm. Data are representative of three independent experiments (A–D, H–J). For transcriptome analysis, biological duplicates were analyzed (E–G). Ordinary one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (A–C, I) or a two-tailed unpaired Student’s t-test for comparing two groups (D, H) (A–D, H, I; error bars, standard deviation [SD] of biological replicates).
Figure 3—figure supplement 1. Characterization of MEK1/2-deficient osteoblast-lineage cells.
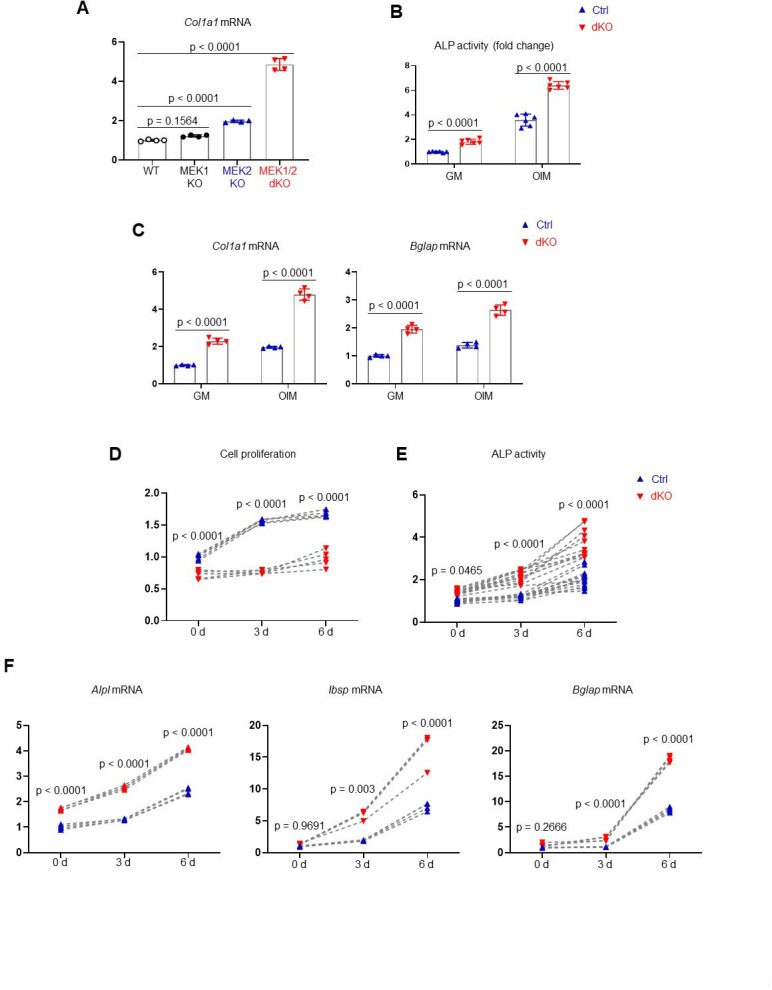
Figure 3—figure supplement 2. Transcriptome analysis of MEK1/2-deficient osteoblast-lineage cells.
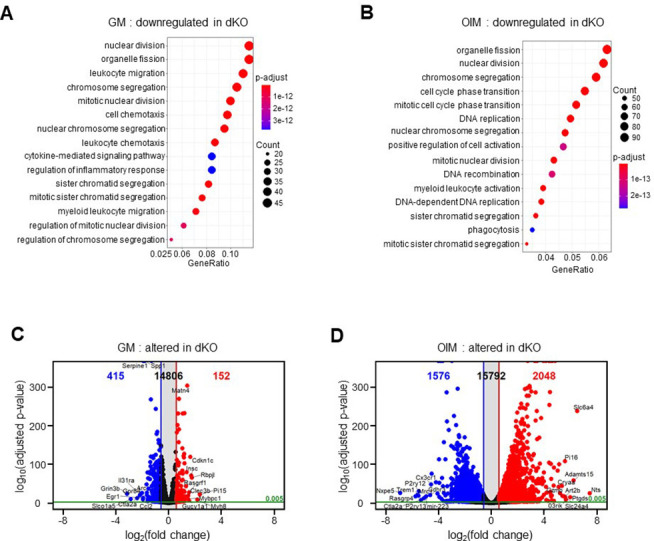
Figure 3—figure supplement 3. MEK1/2-deficient osteoblasts show gene enrichment of WNT and TGF-β signaling.
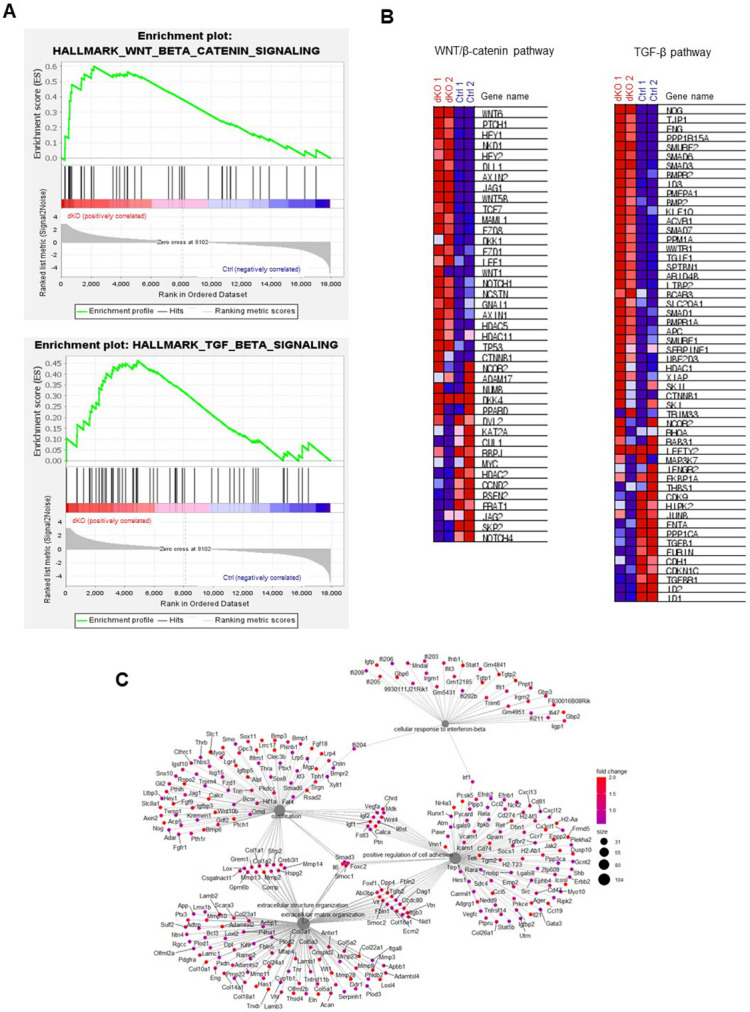
Figure 3—figure supplement 4. MEK1/2-deficient osteoblasts show enhanced WNT/β-catenin and TGF-β signaling at late stages of osteogenic differentiation.
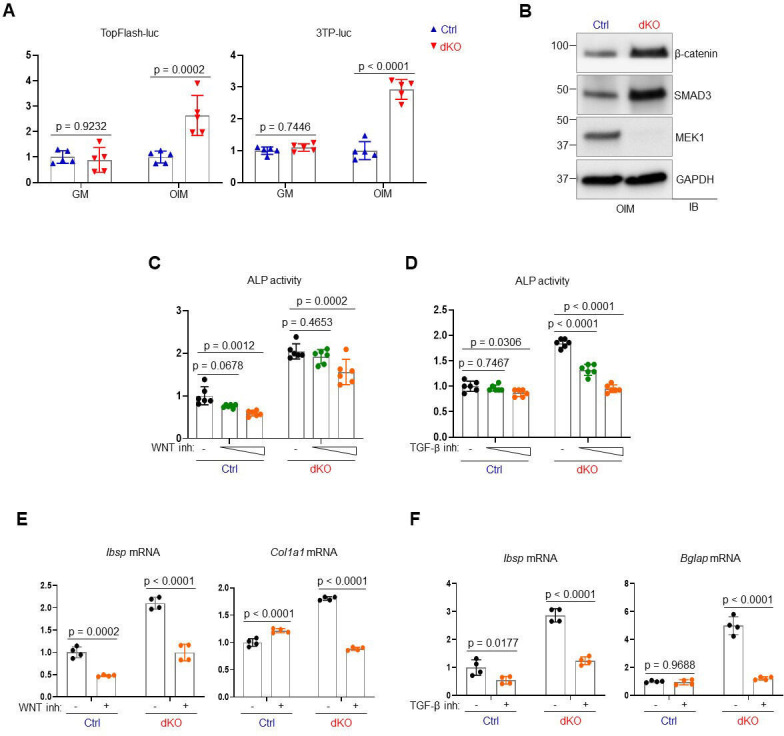
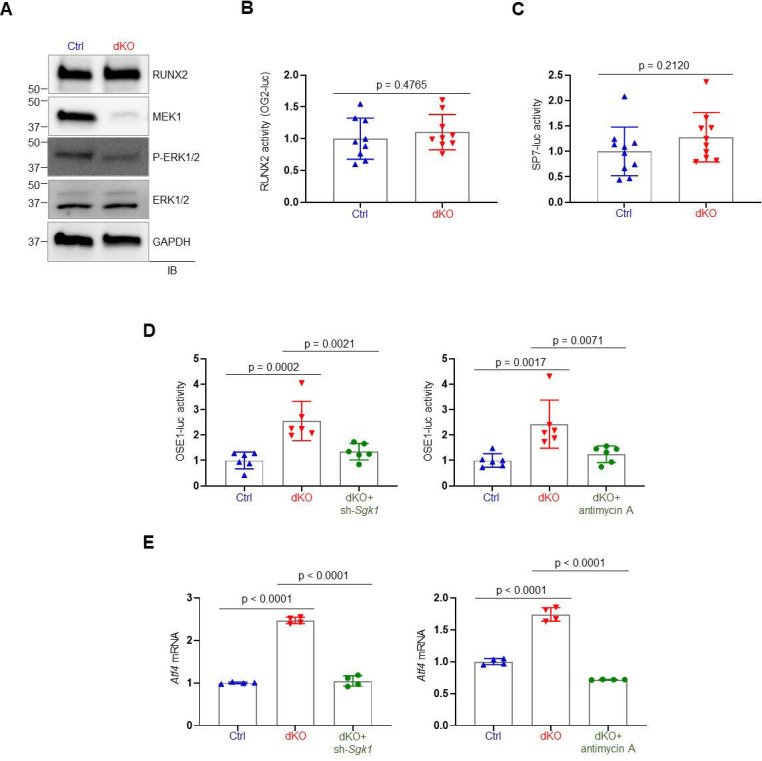
Figure 3—figure supplement 5. Effects of MEK1/2 deletion on RUNX2 expression and transcriptional activity in osteoblasts.