Abstract
Selenium compounds exhibit excellent anticancer properties but have a narrow therapeutic window. Selenium nanoparticles, however, are less toxic compared to other selenium forms, and their biogenic production leads to improved bioavailability. Herein, we used the probiotic strain Lactobacillus casei ATCC 393, previously shown to inhibit colon cancer cell growth, to synthesize biogenic selenium nanoparticles. We examined the anticancer activity of orally administered L. casei, L. casei-derived selenium nanoparticles and selenium nanoparticle-enriched L. casei, and investigated their antitumor potential in the CT26 syngeneic colorectal cancer model in BALB/c mice. Our results indicate that L. casei-derived selenium nanoparticles and selenium nanoparticle-enriched L. casei exert cancer-specific antiproliferative activity in vitro. Moreover, the nanoparticles were found to induce apoptosis and elevate reactive oxygen species levels in cancer cells. It is noteworthy that, when administered orally, selenium nanoparticle-enriched L. casei attenuated the growth of colon carcinoma in mice more effectively than the isolated nanoparticles or L. casei, suggesting a potential additive effect of the nanoparticles and the probiotic. To the best of our knowledge this is the first comparative study examining the anticancer effects of selenium nanoparticles synthesized by a microorganism, the selenium nanoparticle-enriched microorganism and the sole microorganism.
Biogenic selenium nanoparticles synthesized by the probiotic Lactobacillus casei inhibit colon cancer cell growth in vitro as well as in vivo when administered orally in mice.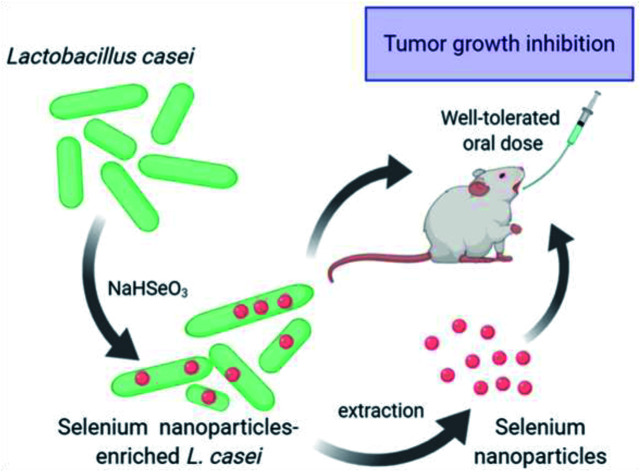
1. Introduction
Selenium (Se) is a nonmetal chemical element primarily found immobilized in sedimentary rocks and soils that has been recognized as an essential trace element.1 Immobilized Se becomes bioavailable through either the weathering of the soil or the reduction by microorganisms.2 It has become evident that Se dietary supplementation has several beneficial effects on human health.3,4 Importantly, Se has been reported as a promising compound in cancer prevention and therapy.5 Moreover, its anticancer properties have been demonstrated by clinical trials where the most pronounced effects of Se-supplementation were observed against colorectal, lung, and prostate cancers.6 There is a narrow window between the beneficial/therapeutic Se dose and the dose that exerts toxicity,7 while the degree and mechanism of its toxicity appear to be strictly dependent on its chemical form.8
Selenium nanoparticles (nano-Se), due to their nanoscale size and lower surface-area-to-volume ratio, possess distinct physicochemical characteristics compared to other Se forms. Besides being less toxic,9 nano-Se have also better bioavailability compared to both Se salts10 and commercially available yeast-based Se-supplements.11 Therefore, nano-Se are considered an excellent candidate for the replacement of other forms of Se in clinical practice.12–14
Today the most common approach for the synthesis of nano-Se is chemical reduction.15 However, chemical synthesis is costly, requires special equipment as well as toxic chemicals. It is noteworthy that, in recent years, there has been a great increase in interest in exploiting biosynthetic routes.16 In particular, various microorganisms have been reported to synthesize nano-Se through their detoxification mechanisms17 or redox homeostasis.18 Among the microorganisms able to resist Se toxicity by biologically converting Se oxyanions to the less toxic Se(0), there are certain bacteria that, in the process of Se-anion reduction, synthesize nanoparticles of elemental Se.19
Se nanoparticles synthesized by bacteria have unique arrangements of Se atoms that vary greatly among different species. These differences reflect the diversity of the enzymes involved in the reduction of Se-oxyanions in different microorganisms.20 The variety of bacteria-derived nano-Se formulations demonstrates the complexity of their physicochemical properties and, eventually, the diversity of their biological effects. As previously stated in Oremland et al.,21 such variety and specificity are difficult to achieve by the traditional methods of synthesis.
Among the various microorganisms that are able to produce nano-Se are Desulfovibrio desulfuricans,18Stenotrophomonas maltophilia, Rhodobacter sphaeroides,22Pseudomonas alcaliphila,23Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei10 and Lactobacillus plantarum,24 the last three being probiotic strains that belong to the genus Lactobacillus.
Enrichment with nano-Se of certain probiotic microorganisms that possess antiproliferative properties was recently shown to enhance their anticancer activity in vivo. In particular, nano-Se enriched Lactobacillus plantarum ATCC 8014 inhibited the growth of 4T1 breast tumors in BALB/c mice24 and nano-Se enriched Lactobacillus brevis extended the life span and inhibited metastasis to liver in the same 4T1 breast tumor model.25
During the last few years, considerable attention has been paid to the beneficial properties of probiotics, especially to the different species of the genus Lactobacillus which have been shown to exert immunomodulatory, anti-inflammatory and anti-carcinogenic strain-specific activities.26–28 Our team has been studying the antiproliferative effects of L. casei ATCC 393 against colon cancer. We have reported that the oral administration of L. casei ATCC 393 significantly impaired tumor growth and raised immune responses in an experimental in vivo colon carcinoma model.28,29
This specific Lactobacillus strain was first reported to accumulate Se intracellularly in 1995 by Calomme et al.30 Even though the researchers did not identify the nanoparticulate nature of these Se aggregates in L. casei ATCC 393, they were the first to propose the application of Se-enriched probiotics in nutritional supplementation studies. Later investigations confirmed the synthesis of Se nanoparticles by various probiotic strains10 and it was only after 2018 that L. casei ATCC 393 was found to be among the nano-Se producing probiotic strains.31
Herein, we used the probiotic strain Lactobacillus casei ATCC 393 (L. casei) to synthesize biogenic nano-Se which we isolated and characterized. Moreover, we comparatively examined the anticancer activity of L. casei, L. casei-derived nano-Se and nano-Se-enriched L. casei, and investigated their efficacy as dietary supplements/nutraceuticals for the suppression of colon cancer cell growth in a syngeneic mouse tumor model. Our results indicate that L. casei-derived nano-Se and nano-Se-enriched L. casei exert cancer-specific antiproliferative activity against colon cancer cells. It is noteworthy that nano-Se-enriched L. casei attenuated the growth of colon carcinoma in mice more effectively than L. casei-derived nano-Se or L. casei, indicating an additive effect between the nanoparticles and the probiotic.
2. Results and discussion
NaHSeO3 as a Se source for Lactobacillus casei
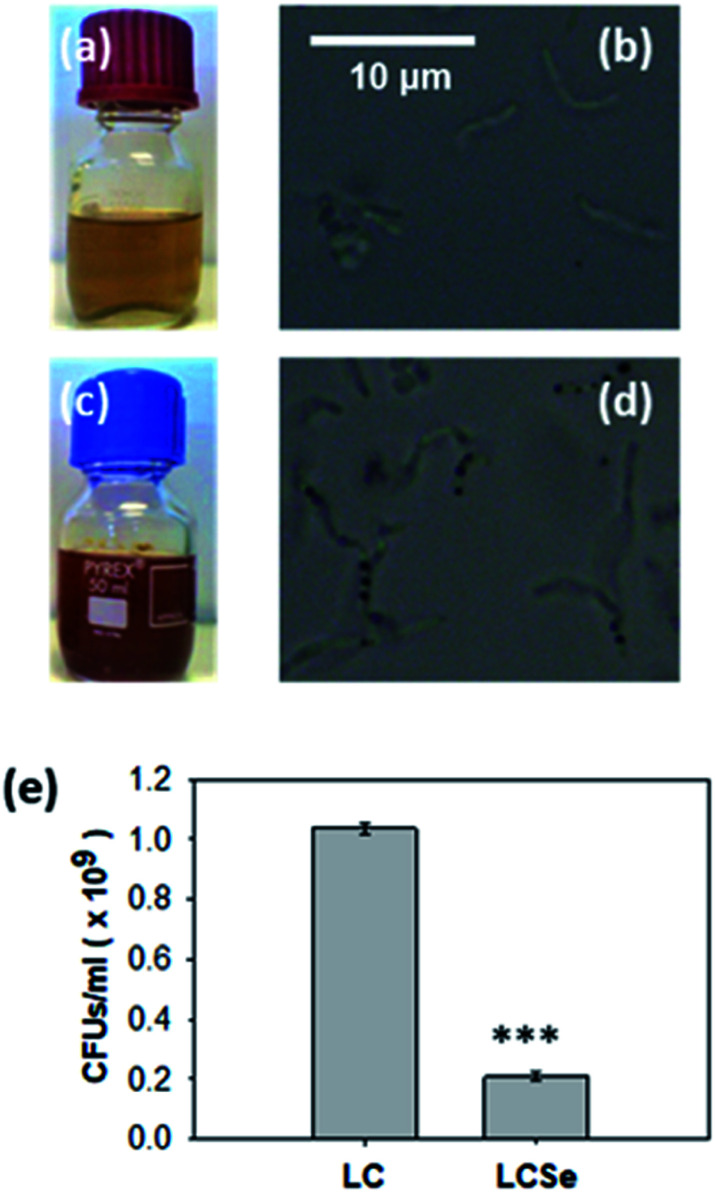
For the biosynthesis of nano-Se by L. casei (LC), bacterial cultures were supplemented with 20 μg ml−1 NaHSeO3 as a Se source and incubated for 96 h until the color of the culture medium turned red (Fig. 1). Interestingly, the bacteria that had grown in the presence of NaHSeO3 (LCSe), when observed under an optical microscope, appeared to have intracellular aggregated sphere-shaped conglomerates (Fig. 1(d)). As expected, bacterial growth was inhibited by NaHSeO3. The exact CFUs per ml of the LC or LCSe cultures were determined by counting the colonies grown on agar plates spread with LC or LCSe bacteria in their late log/early stationary phase of culture and found to be (1.037 ± 0.018) × 109 CFUs per ml for LC and (0.210 ± 0.014) × 109 CFUs per ml for LCSe (Fig. 1(e)).
Fig. 1. Growth of L. casei in the presence of NaHSeO3. Growth of L. casei in (a, b) MRS or (c, d) MRS supplemented with NaHSeO3. Red color of NaHSeO3-supplied culture compared to standard culture indicates the presence of red elemental selenium. Optical microscopy images (magnification 100×) of bacteria grown in (b) MRS or (d) MRS supplemented with NaHSeO3 indicate the presence of intracellular sphere-shaped conglomerates only in NaHSeO3-supplied culture. (e) CFUs in L. casei cultures grown in MRS (LC) or MRS supplemented with NaHSeO3 (LCSe). Both cultures were grown for 19 h with an inoculum of 107 CFUs per ml. CFUs were determined by agar plate counting and three plates per sample were examined. Results presented are representative of three independent experiments. Asterisk indicates a statistically significant difference (Student's t-test, p ≤ 0.001).
Inhibition of LC growth by NaHSeO3 is in agreement with previously reported findings where the growth rate of L. bulgaricus was found to be significantly inhibited in the presence of a Se source in the same concentration range as the one used in our experiments.32 Moreover, the observation that NaHSeO3 delays the L. casei growth demonstrates the fact that lactobacilli might synthesize nano-Se in order to convert the toxic forms of Se to which they are being exposed, in a relatively harmless form.33
Extraction and characterization of Se nanoparticles from Lactobacillus casei
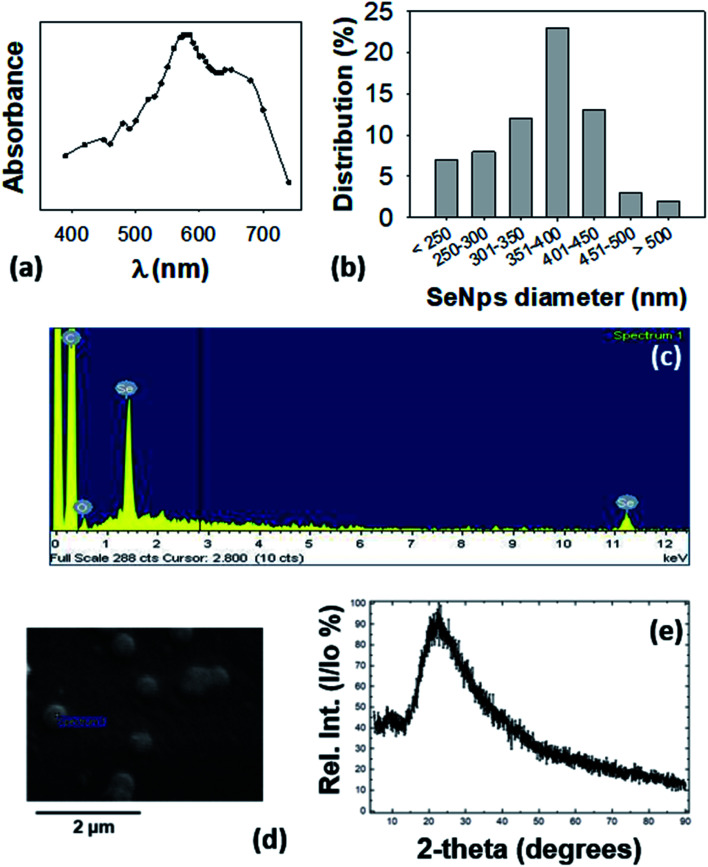
The observed intracellular conglomerates were extracted from LCSe bacteria and purified. The visible spectrum of isolated conglomerates revealed a peak at 575–585 nm as shown in Fig. 2(a) which is consistent with previously reported absorption spectra of nano-Se.34–36 Various species of the genus Lactobacillus were shown to possess the ability to synthesize nano-Se. In particular, both Lactobacillus acidophilus and Lactobacillus helveticus10 produced nano-Se when they were exposed to NaHSeO3. In addition, nano-Se were successfully produced by L. plantarum37 and L. brevis.25 It is noteworthy that L. casei was also found to be able to synthesize nano-Se.10
Fig. 2. Characterization of extracted SeNps. (a) UV-Vis analysis of SeNps extracted and purified from lysed L. casei bacteria cultured in the presence of NaHSeO3, and dispersed in de-ionized water. Note the peak at 575–585 nm. (b) Size distribution of SeNps analyzed by ImageJ analysis of SEM images. SeNps have a diameter of approximately 170–550 nm with an average of 360 nm and a median of 379 nm. (c) Energy dispersive spectroscopy analysis of the SEM images revealing emission peaks of Se and (d) SEM images of purified SeNps. (e) X-ray diffraction pattern of SeNps.
SEM analysis revealed that the LCSe-isolated conglomerates have a spherical shape (Fig. 2(d)) and a diameter in the nanoscale range of approximately 170–550 nm with an average of 360 nm and a median of 380 nm (Fig. 2(b)). Furthermore, the signature peaks of Se were detected in the EDS spectrum (Fig. 2(c)) of the conglomerates; thus, we concluded that the conglomerates are spherical nano-Se (SeNps). Our observation, that the probiotic strain L. casei ATCC 393 is able to synthesize nano-Se, has been further confirmed by three recently published papers.31,38,39
Most of the microorganisms that are able to produce nano-Se synthesize nanoparticles of a spherical shape.40 Their size varies significantly because it depends on the different experimental parameters employed (microbial strain, Se source, incubation period). The polydispersity (170–550 nm, Fig. 2(b)) of the size distribution of the SeNps produced in this work is in good agreement with the literature where the size range of biogenic nano-Se varies between 50 and 500 nm. The average diameter lies at the upper end of the reported distribution range as we observed a Se nanoparticulate formulation of a medium size of 360 nm, while the most common size reported in the literature is ≈100 nm.16,23,38,41
We also characterized the isolated nanoparticles by X-ray diffraction (XRD). Their XRD pattern showed a broad peak at 2θ angles of 15–350° (Fig. 2(e)) which suggests that the nanoparticles are amorphous and not crystalline.16 The amorphous nature of SeNps is also suggested by their red color. Indeed, biogenic reduction of Se oxyanions induces the formation of amorphous spherical nanoparticles.16,21
Conclusively, by following the method described, from 1010 CFUs of LCSe bacteria a nanoparticle yield of 1500 μg of spherical, red amorphous SeNps with a mean diameter of 360 nm was achieved.
Comparative analysis of the in vitro antiproliferative and pro-apoptotic activity of SeNps, LCSe and LC
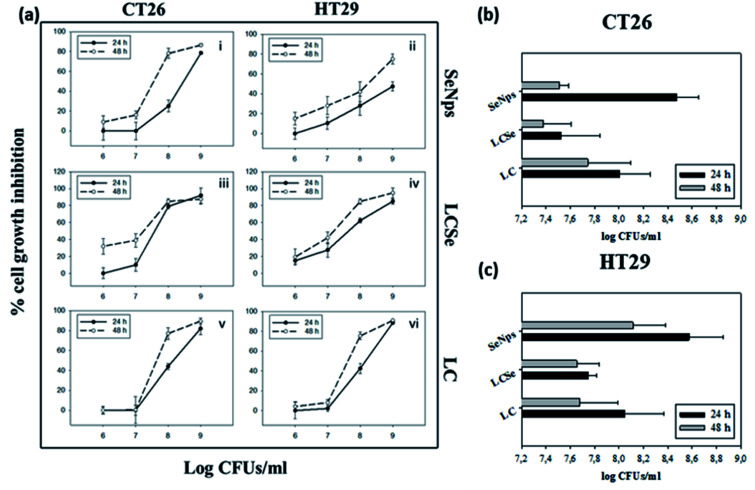
LCSe-isolated SeNps inhibited CT26 and HT29 colon cancer cell growth of murine or human origin, respectively, in a time- and concentration-dependent manner, while a similar effect was observed for LCSe and LC (Fig. 3(a)). The smaller EC50 values against CT26 for 24 or 48 hours estimated for all the treatment groups indicate that CT26 cells are more susceptible to the antiproliferative effects induced by SeNps, LCSe or LC than HT29 cells under these experimental settings. Specifically, the EC50 values for LCSe after 24 h or 48 h were 7.53 or 7.38 CFUs per ml for CT26 cells, compared to 8.00 or 7.74 for LC respectively and 8.47 or 7.51 for SeNps (Fig. 3(b)). For HT29 cells, the EC50 values for LCSe after 24 h or 48 h were 7.74 or 7.65 CFUs per ml compared to 8.05 or 7.68 for LC respectively and 8.57 or 7.51 for SeNps (Fig. 3(c)). Both these cell lines are suitable models for the in vitro investigation of the antiproliferative properties of bioactive compounds against colon cancer and are commonly used together in such preclinical studies. These cells, being of different origin, have different genetic and epigenetic alterations and different mutations.42–44 Therefore, the differences in the EC50 values between CT26 and HT29 cells may reflect their phenotypic and functional variability.
Fig. 3. Growth inhibition of CT26 or HT29 cells by SeNps, L casei-SeNps or L. casei. (a) Cell growth inhibition was analyzed by the SRB assay after 24 or 48 h of treatment. Percentages% of growth inhibition compared to control cells are expressed as mean ± SD. from six replicates. Estimated EC50 values for 24 h or 48 h of treatment in CT26 (b) or HT29 (c) cells. L. casei-SeNps (LCSe) refer to L. casei cultured in the presence of NaHSeO3; SeNps refer to the purified SeNps from the L. casei-SeNps culture; LC refers to L. casei. SeNp concentration refers to the LCSe CFUs, from which the SeNps were extracted. Data presented are representative of at least four independent experiments.
Nano-Se have been reported to exhibit antiproliferative effects against various cancer cell lines such as the human Hep-G2 (ref. 45) and H22 (ref. 46) hepatocarcinoma cell lines, the A375 human melanoma cell line,47 the HeLa human cervical carcinoma cells and the MDA-MB-231 (ref. 48 and 49) and MCF7 (ref. 49) human breast cancer cell lines. Furthermore, biogenic nano-Se derived from L. casei ATCC 393 were previously shown to inhibit the proliferation of the human liver tumor cell line Hep-G2.38,50
Regarding in vitro colon cancer models, nano-Se have been shown to inhibit the proliferation of Caco-2,49,51 HCT-8,46 HCT-116 (ref. 49) and Colo-201 (ref. 52) colon cancer cells. It is noteworthy that HT29, the human colon cancer cell line studied in this work, has been previously reported to be sensitive to nano-Se.53,54
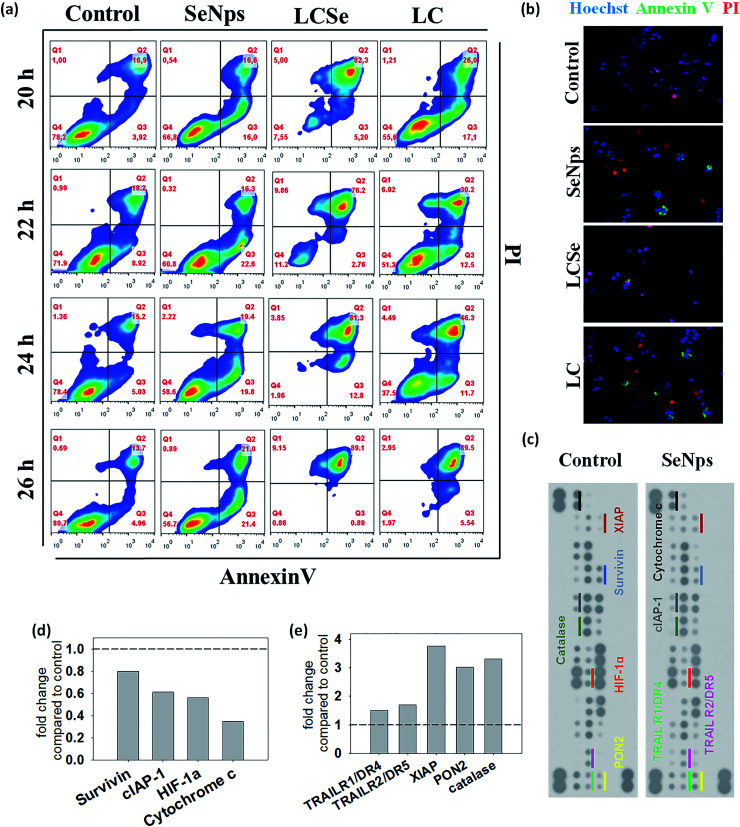
Subsequently, focusing on HT29 colon cancer cells of human origin, we observed that part of the antiproliferative effects exerted by SeNps, LCSe or LC can be attributed to the induction of apoptotic cell death. For the comparative analysis of the apoptotic effects induced by SeNps, LCSe and LC, the time-dependent dynamic process of apoptosis for each treatment group was studied in a pre-defined time frame (20–26 h) for 108 CFUs per ml for LC and LCSe or 15 μg ml−1 for SeNps (15 μg of SeNps were extracted from 108 CFUs of LCSe) by flow cytometric analysis of the double AnnexinV-FITC/PI (AnnexinV-fluorescein isothiocyanate/propidium iodide) staining.
During this time period, all treatments induced apoptosis in HT29 cells, as evident by the early apoptotic cell fraction (AnV + PI−) that moves towards the dead cell fraction (AnV + PI+) (Fig. 4(a)). LC-treated cells move rapidly from the live (AnV− PI−) respectively. As expected, control cells do not present significant alterations in their distribution to the different AnV/PI fractions. The flow cytometry results illustrated in Fig. 4(a) were confirmed with fluorescence microscopy imaging of LC-, LCSe- or SeNp-treated cells under the same conditions for 20 h (Fig. 4(b)).
Fig. 4. Detection of apoptosis induced in HT29 cells. (a) Cells were treated with L. casei (LC), L. casei-SeNps (LCSe) or SeNps for 20–26 h. Cells were stained with Annexin V-FITC and PI, and analyzed by flow cytometry. (b) Fluorescence microscopy images of apoptotic/necrotic cells treated with LC, LCSe or SeNps for 20 h. Cells were stained with Hoechst (blue), Annexin V-FITC (green) and PI (red) and observed under a fluorescence microscope. (c) Expression of apoptosis-related proteins in HT29 cells after SeNp treatment; images of dot plots of protein levels analyzed with a human apoptosis array (ARY009, R&D Systems). Indicated duplicate dots represent a protein expressed in either control or SeNp-treated cells (20 h). Fold change compared to control in the expression levels of selected proteins exhibiting (d) down- or (e) up-regulation upon treatment with SeNps. Dashed lines indicate the expression level in control cells. Cells in all experiments were treated with 108 CFUs per ml of either LC or LCSe, or 15 μg ml−1 of SeNps. The SeNp quantity of 15 μg corresponds to the amount of the nanoparticles extracted from 108 CFUs of LCSe.
Probiotics are known to induce and regulate apoptosis in cancer cells by various mechanisms;55 the pro-apoptotic activity of Lactobacillus casei ATCC 393 has been previously reported by our team.29 Regarding SeNps, it has been suggested that the main mechanism behind the direct anticancer activity of nano-Se is apoptosis.53 Kong et al.56 showed that nano-Se induce caspase-mediated apoptosis in prostate LNCaP cancer cells; Zhang et al.57 showed that nano-Se induce apoptotic cell death in Hep-G2 hepatocarcinoma cells and Yang et al.58 reported the pro-apoptotic effects of nano-Se against the A375 human melanoma cancer cell line. In all the above-mentioned studies nano-Se were synthesized via chemical routes.
Even though there are several studies on the pro-apoptotic effects of chemically synthesized nano-Se or biogenic nano-Se that have been coupled with various therapeutic/targeting compounds,57,59,60 there is a paucity of literature on the pro-apoptotic effects of naked biogenic SeNps. As of today and to the best of our knowledge the pro-apoptotic activity of naked but chemically synthesized SeNps has been reported in MCF-7 (ref. 61) and A375 (ref. 47) cells in vitro and in a hepatocellular carcinoma model in rats in vivo.62
SeNps modulate the protein levels of apoptosis-related molecules and elevate ROS levels in HT29 cells
In order to explore the potential molecular mechanisms of SeNp-induced apoptosis in HT29 cells, the levels of certain apoptosis-related proteins were analyzed in cell lysates. Our results show that various apoptotic signaling proteins were modulated in SeNp-treated HT29 cells (Fig. 4(c)).
The intrinsic apoptosis pathway is inhibited by a group of proteins with structural similarities called IAPs (Inhibitors of Apoptosis Proteins). In humans, there are 6 such proteins, three of which were downregulated by SeNps. Namely as shown in Fig. 4(d), Survivin was downregulated to 0.8-fold and cIAP1 to 0.6-fold. XIAP, the most well studied protein among IAPs, was upregulated by SeNps to 3.8-fold (Fig. 4(e)). Interestingly, besides its caspase-inhibitory and anti-apoptotic role, XIAP is also critically involved in the cellular response to oxidative stress by modulating ROS levels.63
The TRAIL death receptors DR4 and DR5 were also upregulated, both at 1.5-fold as illustrated in Fig. 4(e). These receptors bind TRAIL, a type II and potent apoptosis-inducing transmembrane protein with cancer cell specific action. Interestingly, HT29 cells are TRAIL-resistant,64 so SeNps might induce sensitization of TRAIL-induced apoptosis by upregulating TRAIL death receptors DR4 and DR5.
Two more proteins with anti-oxidant activity were upregulated by SeNps, both exhibiting a 3-fold rise: PON2 and Catalase (Fig. 4(e)). PON2 is a membrane bound protein that prevents ROS formation. It is noteworthy that overexpression of PON2 has been recently described to inhibit cancer development in a mouse model.65 Catalase, an enzyme that protects the cell from oxidative damage,66 has been reported to prevent experimental tumor metastasis of colon cancer cells to the lungs and liver.67 HIF-1a, a subunit of the heterodimeric transcription factor HIF-1 that plays an important role in hypoxia, was downregulated (0.5-fold) by SeNps (Fig. 4(d)). HIF-1a, subunit of the heterodimeric transcription factor HIF-1 that plays an important role in hypoxia and has been a target for the development of anticancer drugs that aim to reverse hypoxic resistance to apoptosis in cancer cells,68 was downregulated (0.5-fold) by SeNp (Fig. 4(d)).
Cytochrome c is one of the central molecules in apoptotic pathways. In response to apoptotic stimuli, it is being released from the mitochondria to the cytosol where it is involved in the activation of caspases.69 In HT29 cells, cytochrome c was downregulated to 0.34-fold by SeNps (Fig. 4(d)). Interestingly, it has been reported that depletion of cytochrome c sensitizes colon cancer cells to a non-apoptotic and non-autophagic death that can be induced by various stimuli. Moreover, it has been shown that downregulation of cytochrome c in HT29 cells sensitizes them in various anticancer compounds. Lastly, it has been described that cytochrome c depletion enhanced the immunogenicity of colon carcinoma cells in a mouse tumor model.70
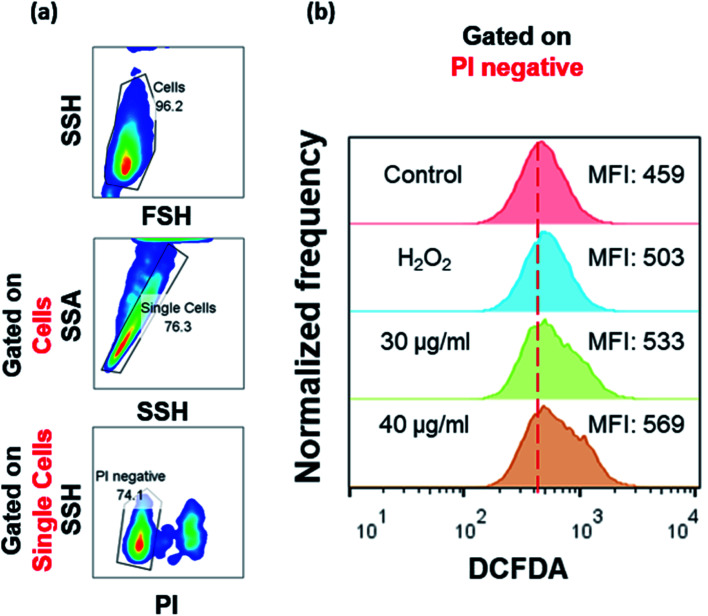
It is noteworthy that nano-Se have been reported to possess both pro-oxidant71 as well as anti-oxidant properties.9 Moreover, it is well documented in the literature that nano-Se, regardless of the method of their synthesis, induce apoptosis by depletion of mitochondrial membrane potential, cytochrome c release and production of ROS.53 Indeed, SeNps were found to induce the formation of ROS in HT29 cells in a concentration dependent manner as evident by the DCFDA (carboxy-H2DCFDA: 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate) flow cytometry histograms in Fig. 5. Our results add to the growing evidence that SeNps induce apoptosis while the observed modulation of the expression levels of various proteins involved in redox regulation and in the intrinsic apoptotic path, and the elevated intracellular ROS levels strengthen the hypothesis that a central mechanism of the SeNp-induced apoptosis might be the induction of ROS generation.
Fig. 5. ROS levels in SeNp-treated HT29 cells. Intracellular levels of ROS were analyzed by flow cytometry using DCFDA. Cells were treated with SeNps (30 or 40 μg ml−1) or 200 μM H2O2 (positive controls) for 24 h. Only live cells (PI negative) were analyzed. (a) Representative flow graphs showing the gating strategy. (b) Representative fluorescence intensity histograms of cells stained with DCFDA from three independent experiments. MFI stands for Median Fluorescence Intensity. Data were analyzed with FlowJo v.10.
SeNps are not toxic to primary murine healthy colonic epithelial cells
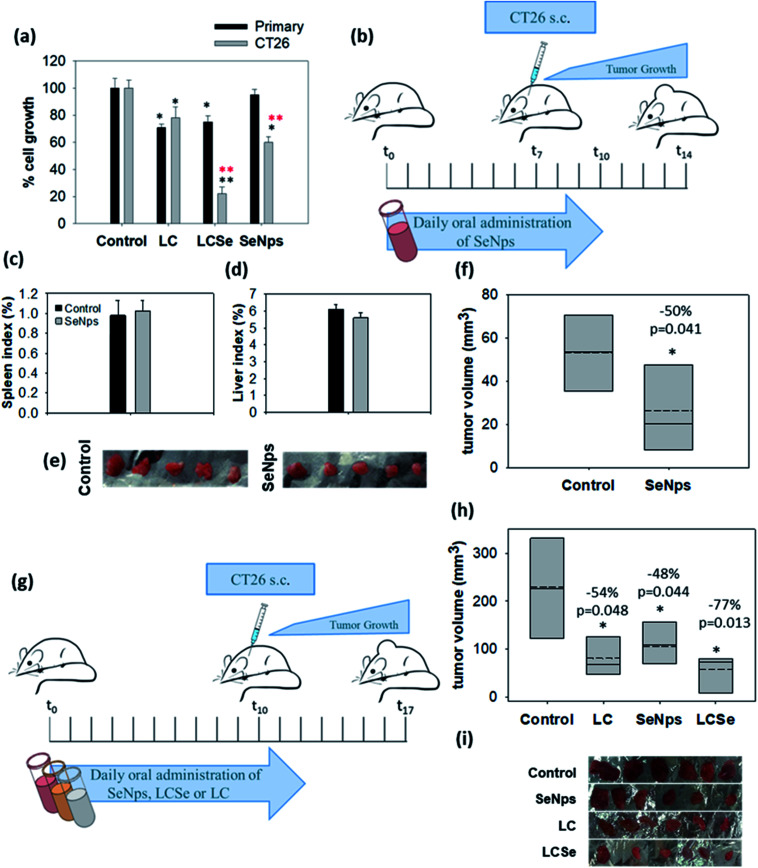
Primary cells were isolated from the colon epithelium of healthy BALB/c mice, cultured and treated with 108 CFUs per ml of LC or LCSe and 15 μg ml−1 of SeNps for 24 h (15 μg of SeNps were extracted from 108 CFUs of LCSe). Contrary to cancer cells, primary healthy cells were not sensitive to SeNps as shown in Fig. 6(a). CT26 cancer cell growth was inhibited by SeNps to 60% compared to control untreated cells, while SeNp-treated primary cells exhibited a similar to control cell growth rate. Moreover, while LCSe inhibited the growth rate of primary cells to 78%, when CT26 cancer cells were exposed to it, their proliferation rate dropped to 25%. On the contrary, under these experimental settings and for the concentrations used LC seemed to exert similar antiproliferative activity against both cancer and primary healthy cells. Thus, contrary to cancer cells, SeNps are harmless and not toxic to primary healthy cells isolated from the colon of BALB/c mice which is in good agreement with the literature where biogenic nano-Se have been reported not to affect the viability of human peripheral blood mononuclear cells isolated from healthy volunteers.72 Moreover there have been previous reports indicating the cancer specificity of the toxic effects exerted by Se compounds.73
Fig. 6. Biogenic SeNps attenuate tumor growth in mice. (a) Growth inhibitory effects in murine cancer CT26 or healthy primary colon cells. Primary cells were isolated from the colon of healthy BALB/c mice. Comparative examination with the SRB assay of the antiproliferative activity of L. casei (LC), L. casei-SeNps (LCSe) and SeNps against either CT26 murine colon cancer cells or primary epithelial colon cells. Cells were treated with 108 CFUs per ml of LC or LCSe, or 15 μg ml−1 of SeNps for 24 h. The quantity of 15 μg of SeNps corresponds to the amount of nanoparticles extracted from 108 CFUs of LCSe. Black asterisks indicate a statistically significant difference compared to control and red asterisks a significant difference between cancer and primary healthy cells. (b) Oral administration of SeNps in mice. Administration scheme. Ten male BALB/c mice were assigned to two groups (n = 5). The SeNps group received daily 150 μg per mouse of SeNps for 10 days, while the Control group received PBS. On the seventh day mice were inoculated with CT26 cells and seven days later they were euthanized. Comparison of mean (c) spleen and (d) liver index between SeNp-treated and Control mice. No significant difference was observed between groups. (e) Photographic observation of tumors excised from SeNp-treated and Control mice and (f) mean volume of the excised tumors. A statistically significant −50% volume inhibition was observed in tumors excised from SeNp-treated mice. (g) Comparative analysis of the effect of SeNPs, LC, and LCSe on tumor growth in mice. Twenty four male BALB/c mice (n = 6) received orally in a prophylactic scheme 109 CFUs of LC or LCSe or 150 μg of SeNps per mouse, once per day, for twelve days. Control mice received PBS. On the tenth day mice were inoculated with 5 × 106 CT26 cells subcutaneously. Mice were euthanized seven days later. (h) Mean volume and (i) photographic observation of tumors excised from SeNps, LC or LCSe-treated and Control mice. A statistically significant reduction in tumor volume was observed in all treatment groups. Statistically significant differences are indicated with asterisks (*p < 0.05, Student's t-test or one-way ANOVA).
These results indicate that both SeNps and LCSe could possess enhanced cancer-specific antiproliferative properties. Considering the narrow therapeutic window of Se,74 this observation is extremely important since it indicates that SeNps could facilitate the therapeutic effects of Se, minimizing its side effects by allowing increased dosage schemes.
Orally administered SeNps inhibit in vivo growth of colon carcinoma
Prophylactic oral administration of SeNps (6.5 mg SeNps per kg of total animal weight or ≈150 μg per mice) in mice for 10 days, as it is described in Fig. 6(b), was well tolerated. Monitoring of mouse weight throughout the experiment did not reveal any significant change between SeNp-treated and Control groups of mice. Moreover, spleen and liver weights were not significantly different between groups (liver index was 6.12% ± 0.29 in Control and 5.6% ± 0.32 in SeNp-treated mice, while the spleen index was 0.98% ± 0.15 in Control and 1.02% ± 0.11 in SeNp-treated mice) (Fig. 6(c) and (d)). Thus, no indication of toxicity was noted, suggesting that the daily administered dose of SeNps is well tolerated. This is in good agreement with Shakibaie et al.75 who studied the toxicity of biogenic, spherical and amorphous nano-Se produced by Bacillus sp. MSh-1. Specifically, the LD50 value of orally administered nano-Se in mice was determined to be 198.1 mg kg−1, which corresponds to a 30-fold higher dose than the one used in our experiments. Moreover, in the same study it was described that daily oral administration of a dose of up to 10 mg kg−1 of biogenic nano-Se was harmless without side effects in mice. Similar to Shakibaie et al.,75 our observations substantiate the well-tolerated nature of biogenic SeNps.
It is noteworthy that the oral administration of SeNps according to the schedule described above (Fig. 6(b)) significantly inhibited CT26 tumor growth in BALB/c mice. Specifically, tumors excised from mice that received SeNps were 50% smaller (p = 0.041, Student's t-test) compared to control animals (Fig. 6(e) and (f)).
Thus, under the above-mentioned experimental settings the orally administered biogenic SeNps produced by LC are well tolerated in mice and induce tumor growth inhibition.
Oral administration of LCSe inhibits the growth of colon carcinoma in mice more effectively than SeNps or LC
Next, we comparatively examined in vivo the effect induced by oral administration of SeNps with LCSe or LC in tumor-bearing BALB/c mice, as described in Fig. 6(g). All treatments (SeNps, LCSe, and LC) induced a statistically significant inhibition in tumor growth (Fig. 6(h) and (i)), but LCSe, in accordance with our in vitro results illustrated in Fig. (3), exhibited the most potent growth-inhibitory activity. Specifically, LCSe inhibited tumor volume by 77% compared to control, while SeNps induced a reduction of −48% and LC of −54% (Fig. 6(h)).
Regarding LC, our results are in good agreement with previous studies describing the growth inhibitory effects of L. casei against colon cancer in vivo.28,29,76,77 Regarding Se-enriched probiotics like LCSe, there is evidence that they can inhibit tumor growth in vivo in pre-clinical studies. In particular, Se-enriched Lactobacillus plantarum ATCC 8014 was shown to inhibit the growth of 4T1 breast tumors in BALB/c mice24 and Se-enriched Bifidobacterium longum successfully suppressed the proliferation of H22 liver tumor cells in Kunming mice.78 Despite the promising results, as of today there are no comparative studies examining the antitumor activity of Se-enriched probiotics. Thus, our observations are extremely important since these findings suggest that the reported growth inhibitory and even pro-apoptotic effects of probiotics may be enhanced via their enrichment with nano-Se.
3. Experimental
Materials, animals and cell lines
Materials
Dulbecco's Modified Eagle's Medium (DMEM) and Fungizone (Amphotericin B) were purchased from Gibco (Gaithersburg, MD, USA). Fetal bovine serum (FBS), trypsin, penicillin/streptomycin, and phosphate-buffered saline (PBS) were purchased from Biosera (Boussens, France). MRS Broth, LAB094 and Agar No. 2 (MC006) were purchased from LabM (UK). The Annexin V-PI kit was purchased from BD Biosciences (USA). The Human Apoptosis Antibody Array kit (ARY 009) from R&D systems (USA) was used. Carboxy-H2DCFDA (C400) was purchased from Invitrogen (Invitrogen, USA). Acetic acid, trichloroacetic acid (TCA), Trizma base, NaHSeO3, H2O2 and sulforhodamine B (SRB) were purchased from Sigma-Aldrich (Germany).
Mammalian cell lines
The colon carcinoma CT26 (murine) and HT29 (human) cell lines were used. CT26 cells are well characterized and consist one of the most commonly used cell lines in drug development.42 HT29 cells are commonly used as an in vitro model for the study of new therapeutic interventions against colon cancer. Both cell lines were cultured under sterile conditions at 37 °C in a humidified incubator with 5% CO2, in DMEM supplemented with 10% fetal bovine serum, penicillin (100 U mL−1) and streptomycin (100 μg mL−1), and 2 mM glutamine.
Primary intestinal (colon) epithelial cells
Primary intestinal epithelial cells were isolated from the colon of three male BALB/c mice (in three individual experiments) as it was previously described.79 The protocol described in Chopra et al. for the rat colon was adapted for the mouse colon. For culturing, 106 isolated cells were seeded in a 60 mm cell culture dish in 3 ml of DMEM supplemented with 15% fetal bovine serum, penicillin (100 units per ml), streptomycin (100 μg ml−1), and Fungizone (2.5 μg ml−1).
Bacteria
The Lactobacillus casei ATCC 393 strain was used for the synthesis of SeNps. L. casei ATCC 393 (LC) was grown in MRS broth in 10 ml glass test tubes without agitation at 37 °C. For subculturing 108 CFUs of LC were inoculated in 10 ml MRS. The cultures reached their late-log/early stationary phase (109 CFUs per ml) 19 h post the inoculation. Bacteria were harvested from late-log/early stationary phase cultures by centrifugation at 1700g for 15 minutes at 4 °C. After being washed with PBS, bacteria were resuspended in either DMEM (for the in vitro experiments) or PBS (for the in vivo experiments) at the indicated CFUs per ml. For the preparation of Se-enriched LC, MRS broth was supplied with 20–60 μg ml−1 NaHSeO3 inoculated with 107 CFUs per ml of LC, and bacteria were grown for 96 h until reaching the late-log/early stationary phase (LCSe).
Animals
Forty-three male BALB/c mice (6–8 weeks old, weight 20–25 g) were raised in the Animal house unit (Laboratory of Experimental Surgery and Surgical Research at Democritus University of Thrace). Mice were kept in polycarbonate cages and provided with tap water ad libitum and a commercial pelleted diet (Mucedola). Animal experiments were approved by the Animal Care and Use Committee of the Veterinary Department of Evros Prefecture since it complied with the requirements set by Directive 86/609/EEC and PD 160/91. All animal experiments were conducted in accordance with the 3 R's (replacement, refinement, and reduction).
Methods
Colony forming units
For the determination of colony-forming units (CFUs) in the bacterial cultures, aliquots of a late log/early stationary LC or LCSe culture were used for seeding the agar plates. For the preparation of the MRS Agar, the LAB094 MRS Broth and LAB MC006 Agar No. 2 were used. 100 μl of samples were seeded per 100 mm-petri plate filled with 10 ml of MRS Agar. Plates were incubated at 37 °C for 70 h in an anaerobic incubator. CFUs were manually counted on the plates seeded with the 100 000-fold dilution of the initial culture.
Extraction of SeNps from Lactobacillus casei
The method for the isolation of SeNps from bacteria is an adaptation from the protocol proposed in Kurek et al.80 First, LCSe bacteria were harvested from the MRS broth by centrifugation (1700×g, 15 min, 4 °C). Following three washes with PBS, bacteria were resuspended in NaOH 0.1 M (5 ml of NaOH was used per 0.1 g of bacteria) and incubated at 37 °C for 90 min. The mixture of lysed bacteria and SeNp-clusters was recovered by five subsequent washes with de-ionized water (7000×g, 15 min, 4 °C). Harvested clusters containing SeNps were sonicated (20 rounds of a 5 seconds sonication period, 70% amplitude, 50 W) using the UP50H sonifier from Hielscher Ultrasonics GmbH (Teltow, Germany). Lastly, the dispersed SeNps were purified from cell debris by vacuum filtration using membrane filters with the pore size ranging from 2 to 0.45 μm. The purified aqueous solution of SeNps was stored at 4 °C.
UV-Vis spectroscopy of SeNps
We analyzed the absorption spectrum of the purified SeNps using the Enspire multi-plate reader (Perkin Elmer). Briefly, 100 μl of SeNps suspended in deionized water were seeded in a 96-well-plate. Readings were normalized to blank wells containing deionized water.
Scanning electron microscopy and energy dispersion spectroscopy
Scanning electron microscopy (SEM) and Energy Dispersion Spectroscopy (EDS) of extracted SeNps were carried out using a JEOL JSM-6390LV scanning microscope equipped with an energy-dispersive X-ray (EDS) INCA micro-analytical system. Operating conditions were: accelerating voltage 20 kV, probe current 45 nA and counting time 60 s, with ZAF correction being provided on-line. The samples were coated with carbon, using a Jeol JEE-4X vacuum evaporator. Size distribution of the nanoparticles was determined by ImageJ analysis of multiple SEM images. Results presented are representative of three independent experiments.
Characterization of SeNps by X-ray crystallography
The structural characterization of SeNps was carried out using XRD diffractograms with Cu-Kα radiation (λ = 1.5405 Å) in a Bragg–Brentano geometry and analyzed with the Rietveld method.
Cell viability assay
Cell viability was determined by the SRB assay.81 CT26 or HT29 or primary murine colon epithelial cells were seeded in 96-well plates at a cell density of 5000 cells per well for CT26 or 20 000 for HT29 or primary cells. After treatment, the cells were fixed using ice-cold 10% TCA for 1 h at 4 °C, washed with tap water and dried before being stained with SRB (0.057% w/v) for 30 minutes at room temperature. The excess dye was removed with 1% acetic acid and the cells were dried. Finally, the protein-bound dye was dissolved in 10 mM Tris base and OD (optical density) was measured at 492 nm using the Enspire multi-plate reader (Perkin Elmer). Background levels of OD due to the presence of LC, LCSe or SeNps were excluded. The % inhibition of cell growth was calculated by eqn (1):
| % growth inhibition = 100 − [(mean OD sample)/(mean OD control) × 100] | 1 |
For the estimation of EC50 values, regression analysis via the four-parameter logistic curve was performed using the Sigma plot software v.11.
Light microscopy
For the observation of the bacteria under a light microscope, a Zeiss PrimoVert Inverted Microscope equipped with a ZEISS Axiocam ERC 5C camera was used.
Analysis of apoptosis with the Annexin V-PI assay
Apoptosis was assayed using the Annexin V-PI double staining assay. A commercially available kit (BD Biosciences) was used according to the instructions. HT29 cells were seeded in 10 cm2 cell culture plates at a density of 2 × 106 cells per plate. Upon reaching 70–80% confluency, the cells were treated with LC, LCSe or SeNps as indicated. Non-treated cells were used as control. Following treatment, the cells were collected by trypsinization and resuspended in Annexin binding buffer. The cells were labeled by 15 minutes-incubation in the dark at room temperature, with Annexin V-FITC and PI and then analysed by flow cytometry (Attune NxT, Thermo Fisher Scientific). Data analysis was performed with FlowJo V10 software.
Intracellular ROS levels
ROS levels were detected by flow cytometry using carboxy-H2DCFDA. Briefly, 3 × 106 HT29 cells per well were seeded in 6-well-plates and left to adhere overnight. The cells were treated with SeNps (30 or 40 μg ml−1) or 200 μM H2O2 (positive control) for 24 h. Control untreated cells were also included in the experiment. After treatment, the cells were collected with trypsinization and incubated in 18 μM DCFDA in PBS at 37 °C for 40 minutes. The cells were washed and additionally labeled with 2.5 μg ml−1 PI for the discrimination of dead cells right before being analyzed with the flow cytometer (Attune NxT, Thermo Fisher Scientific). The cells were excluded based on the PI signal (only PI-negative, thus live cells were analyzed). Data were analyzed with FlowJo v.10.
Oral administration of SeNps in the CT26 mouse tumor model
Twenty male mice were assigned to two groups (n = 10), SeNps and Control. ‘Control’ animals received daily 150 μl of PBS per os while animals in the ‘SeNps’ group received 150 μg of SeNps suspended in 150 μl of PBS. The SeNps dose of 150 μg corresponds to the amount of the nanoparticles extracted from 109 CFUs of LCSe. Oral administration was performed using a gavage needle. On the seventh day, all mice were inoculated with 5 × 106 CT26 cells subcutaneously in the scuff of the neck, while the oral administration of SeNps or PBS was continued for the following 3 days. Seven days post CT26 inoculation, mice were euthanized by cervical dislocation and tumors were excised. Tumor dimensions were measured using an electronic micrometer and tumor volume was calculated using the modified ellipsoid formula (eqn (2))
| [(width2 × length)/2] | 2 |
During the course of the experiments the weight change of each mouse was recorded and all mice were monitored for signs of disease or discomfort. The spleen and liver were also harvested and weighed. The respective liver and spleen% indexes were calculated with the following formula (eqn (3)):
| ([(spleen/liver weight)/body weight] × 100%) | 3 |
Oral administration of SeNps, L. casei-SeNps and L. casei in the CT26 mouse tumor model
For the comparative evaluation of the in vivo growth inhibitory effects of SeNps, LC or LCSe employing the CT26 tumor model, a previously described experimental protocol was followed.28,29,82–84 Briefly, 24 male mice were randomly assigned to 4 groups. Mice in each group received a daily dose of 109 CFUs of either LC or LCSe, or 150 μg of SeNps. The nanoparticles or the bacteria were suspended in 150 μl of PBS and were administered via a gavage needle for 12 days. The SeNps dose of 150 μg corresponds to the amount of the nanoparticles extracted from 109 CFUs of LCSe. At day ten, 5 × 106 CT26 cells per mouse were injected subcutaneously and the protocol described in the previous paragraph was followed.
Data analysis and statistics
All data are representative of at least three independent experiments and the values are presented as mean ± SD. All data were analyzed with Sigma Plot v. 11.0. Statistical comparisons between groups were performed using the Student's t-test or one-way ANOVA where appropriate. Differences between groups were considered statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
4. Conclusions
This study provides evidence that SeNps produced by L. casei and SeNp-enriched L. casei could be good candidates for the development of oral formulations or dietary supplements for the chemoprevention of colon cancer. The evidence presented here indicates that SeNp-enriched probiotic bacteria exert the most potent growth inhibitory effect in vitro and in vivo as compared to either the isolated SeNps or the sole probiotic, and underlines the importance of comparatively evaluating the biological effects of nanoparticle-enriched probiotics. Moreover, our study adds to the growing evidence that SeNps induce apoptosis in colon cancer cells, possible by inducing ROS generation, among other mechanisms. Along with their strong anticancer potential, the isolated SeNps were also demonstrated to have biocompatibility and cancer-specific growth inhibitory action. However, the anti-proliferative and pro-apoptotic effects exerted by the SeNps and the SeNp-enriched bacteria need to be further characterized and the molecular pathways involved need to be identified. Besides colon cancer, the antiproliferative effects of the SeNps should be investigated against other tumor types in different administration schemes.
While our findings cannot be generalized for various types of cancer or different lactobacilli species, they have revealed the strong potential of nanoparticle-enriched probiotics for cancer prevention while highlighting the importance of comparative assessment and exploitation of their bioactivities. We suggest that future studies on biogenic nanoparticles utilize comparative experimental settings with the aim of figuring out the complex interactions between bacteria and bacteria-derived nanoparticles that add up to their highly unique bioactivities.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors would like to thank Assoc. Prof. Dr Yannis Kourkoutas (Laboratory of Applied Microbiology & Biotechnology, Dept. of Molecular Biology and Genetics, DUTh) for the Lactobacillus casei ATCC 393 strain and Prof. Dr Eleni Pavlidou (Electron Microscopy and Structural Characterization of Materials Laboratory, Dept. of Solid State Physics, AUTh) for her help in the SEM and EDS analysis of the nanoparticles. Part of the work was implemented by utilizing the facilities of the project “An Open-Access Research Infrastructure of Chemical Biology and Target-Based Screening Technologies for Human and Animal Health, Agriculture and the Environment (OPENSCREEN-GR)” (MIS 5002691) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund). Part of this study was co-financed by Greece and the European Union (European Social Fund-ESF) through the Operational Programme ‘Human Resources Development, Education and Lifelong Learning 2014-2020’ in the context of the project ‘Biosynthesis of SeNp in probiotic lactobacilli and investigation of their anticancer and immunostimulatory activity’ (MIS 5050556).
References
- Schwarz K. Foltz C. M. J. Am. Chem. Soc. 1957;79:3292–3293. doi: 10.1021/ja01569a087. [DOI] [Google Scholar]
- Sarkar B. Bhattacharjee S. Daware A. Tribedi P. Krishnani K. K. Minhas P. S. Nanoscale Res. Lett. 2015;10:371. doi: 10.1186/s11671-015-1073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman M. P. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- Hosnedlova B. Kepinska M. Skalickova S. Fernandez C. Ruttkay-Nedecky B. Malevu T. D. Sochor J. Baron M. Melcova M. Zidkova J. Kizek R. Int. J. Mol. Sci. 2017;18:2209. doi: 10.3390/ijms18102209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín C. Plano D. Sharma A. K. Palop J. A. Int. J. Mol. Sci. 2012;13:9649–9672. doi: 10.3390/ijms13089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnstedt M. Fernandes A. P. EPMA J. 2010;1:389–395. doi: 10.1007/s13167-010-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel J. K. Power R. Toborek M. IUBMB Life. 2016;68:97–105. doi: 10.1002/iub.1466. [DOI] [PubMed] [Google Scholar]
- Nuttall K. L. Ann. Clin. Lab. Sci. 2006;36:409–420. [PubMed] [Google Scholar]
- Khurana A. Tekula S. Saifi M. A. Venkatesh P. Godugu C. Biomed. Pharmacother. 2019;111:802–812. doi: 10.1016/j.biopha.2018.12.146. [DOI] [PubMed] [Google Scholar]
- Eszenyi P. Sztrik A. Babka B. Prokisch J. Int. J. Biosci., Biochem. Bioinf. 2011;1:148–152. [Google Scholar]
- Hosnedlova B. Kepinska M. Skalickova S. Fernandez C. Ruttkay-Nedecky B. Peng Q. Baron M. Melcova M. Opatrilova R. Zidkova J. Bjørklund G. Sochor J. Kizek R. Int. J. Nanomed. 2018;13:2107–2128. doi: 10.2147/IJN.S157541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalickova S. Milosavljevic V. Cihalova K. Horky P. Richtera L. Adam V. Nutrition. 2017;33:83–90. doi: 10.1016/j.nut.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Zhang J. Wang X. Xu T. Toxicol. Sci. 2008;101:22–31. doi: 10.1093/toxsci/kfm221. [DOI] [PubMed] [Google Scholar]
- Yazdi M. H. Masoudifar M. Varastehmoradi B. Mohammadi E. Kheradmand E. Homayouni S. Shahverdi A. R. Avicenna J. Med. Biotechnol. 2013;5:158–167. [PMC free article] [PubMed] [Google Scholar]
- Shoeibi S. Mozdziak P. Golkar-Narenji A. Top. Curr. Chem. 2017;375:88. doi: 10.1007/s41061-017-0176-x. [DOI] [PubMed] [Google Scholar]
- Husen A. Siddiqi K. S. J. Nanobiotechnol. 2014;12:28. doi: 10.1186/s12951-014-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N. T. Sharma N. Prakash R. Raina K. K. Fellowes J. Pearce C. I. Lloyd J. R. Pattrick R. A. D. Biotechnol. Lett. 2009;31:1857–1862. doi: 10.1007/s10529-009-0096-0. [DOI] [PubMed] [Google Scholar]
- Kessi J. Ramuz M. Wehrli E. Spycher M. Bachofen R. Appl. Environ. Microbiol. 1999;65:4734–4740. doi: 10.1128/AEM.65.11.4734-4740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J. F. Basu P. Santini J. M. Oremland R. S. Annu. Rev. Microbiol. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- Tugarova A. V. Kamnev A. A. Talanta. 2017;174:539–547. doi: 10.1016/j.talanta.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Oremland R. S. Herbel M. J. Blum J. S. Langley S. Beveridge T. J. Ajayan P. M. Sutto T. Ellis A. V. Curran S. Appl. Environ. Microbiol. 2004;70:52–60. doi: 10.1128/AEM.70.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A. Miyashita M. Inoue K. Matsunaga T. Appl. Microbiol. Biotechnol. 1997;48:367–372. doi: 10.1007/s002530051064. [DOI] [PubMed] [Google Scholar]
- Zhang W. Chen Z. Liu H. Zhang L. Gao P. Li D. Colloids Surf., B. 2011;88:196–201. doi: 10.1016/j.colsurfb.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Yazdi M. H. Mahdavi M. Kheradmand E. Shahverdi a R. Arzneimittelforschung. 2012;62:525–531. doi: 10.1055/s-0032-1323700. [DOI] [PubMed] [Google Scholar]
- Yazdi M. H. Mahdavi M. Setayesh N. Esfandyar M. Shahverdi A. R. Daru, J. Pharm. Sci. 2013;21:33. doi: 10.1186/2008-2231-21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de LeBlanc A. de M. LeBlanc J. G. World J. Gastroenterol. 2014;20:16518. doi: 10.3748/wjg.v20.i44.16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco M. L. Pavan S. Kleerebezem M. Curr. Opin. Biotechnol. 2006;17:204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Aindelis G. Tiptiri-Kourpeti A. Lampri E. Spyridopoulou K. Lamprianidou E. Kotsianidis I. Ypsilantis P. Pappa A. Chlichlia K. Cancers. 2020;12:1–16. doi: 10.3390/cancers12020368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiptiri-Kourpeti A. Spyridopoulou K. Santarmaki V. Aindelis G. Tompoulidou E. Lamprianidou E. E. Saxami G. Ypsilantis P. Lampri E. S. Simopoulos C. Kotsianidis I. Galanis A. Kourkoutas Y. Dimitrellou D. Chlichlia K. PLoS One. 2016;11:e0147960. doi: 10.1371/journal.pone.0147960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calomme M. Hu J. Van den Branden K. a Vanden Berghe D. Biol. Trace Elem. Res. 1995;47:379–383. doi: 10.1007/BF02790140. [DOI] [PubMed] [Google Scholar]
- Xu C. Qiao L. Guo Y. Ma L. Cheng Y. Carbohydr. Polym. 2018;195:576–585. doi: 10.1016/j.carbpol.2018.04.110. [DOI] [PubMed] [Google Scholar]
- Palomo-Siguero M. Gutiérrez A. M. Pérez-Conde C. Madrid Y. Microchem. J. 2016;126:488–495. doi: 10.1016/j.microc.2016.01.010. [DOI] [Google Scholar]
- Combs G. F. Garbisu C. Yee B. C. Yee A. Carlson D. E. Smith N. R. Magyarosy A. C. Leighton T. Buchanan B. B. Biol. Trace Elem. Res. 1996;52:209–225. doi: 10.1007/BF02789163. [DOI] [PubMed] [Google Scholar]
- Yu B. You P. Song M. Zhou Y. Yu F. Zheng W. New J. Chem. 2016;40:1118–1123. doi: 10.1039/C5NJ02519B. [DOI] [Google Scholar]
- Li Q. Chen T. Yang F. Liu J. Zheng W. Mater. Lett. 2010;64:614–617. doi: 10.1016/j.matlet.2009.12.019. [DOI] [Google Scholar]
- Khalid A. Tran P. A. Norello R. Simpson D. A. O'Connor A. J. Tomljenovic-Hanic S. Nanoscale. 2016;8:3376–3385. doi: 10.1039/C5NR08771F. [DOI] [PubMed] [Google Scholar]
- Yazdi M. H. Mahdavi M. Varastehmoradi B. Faramarzi M. A. Shahverdi A. R. Biol. Trace Elem. Res. 2012;149:22–28. doi: 10.1007/s12011-012-9402-0. [DOI] [PubMed] [Google Scholar]
- Xu C. Guo Y. Qiao L. Ma L. Cheng Y. Roman A. Front. Microbiol. 2018;9:1129. doi: 10.3389/fmicb.2018.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L. Dou X. Yan S. Zhang B. Xu C. Food Funct. 2020;11:3020–3031. doi: 10.1039/D0FO00132E. [DOI] [PubMed] [Google Scholar]
- Jain R. Jordan N. Weiss S. Foerstendorf H. Heim K. Kacker R. Hübner R. Kramer H. van Hullebusch E. D. Farges F. Lens P. N. L. Environ. Sci. Technol. 2015;49:1713–1720. doi: 10.1021/es5043063. [DOI] [PubMed] [Google Scholar]
- Dhanjal S. Cameotra S. S. Microb. Cell Fact. 2010;9:1–11. doi: 10.1186/1475-2859-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle J. C. Loewer M. Boegel S. de Graaf J. Bender C. Tadmor A. D. Boisguerin V. Bukur T. Sorn P. Paret C. Diken M. Kreiter S. Türeci Ö. Sahin U. BMC Genomics. 2014;15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Bodmer W. F. Proc. Natl. Acad. Sci. U. S. A. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer D. L. Kline T. McCabe F. L. Faucette L. F. Feild J. Chaikin M. Anzano M. Reiman D. Hoffstein S. -J Li D. Gennaro D. Buscarino C. Lynch M. Poste G. Greig R. Int. J. Cancer. 1988;41:287–296. doi: 10.1002/ijc.2910410221. [DOI] [PubMed] [Google Scholar]
- Estevez H. Garcia-Lidon J. C. Luque-Garcia J. L. Camara C. Colloids Surf., B. 2014;122:184–193. doi: 10.1016/j.colsurfb.2014.06.062. [DOI] [PubMed] [Google Scholar]
- Gao F. Yuan Q. Gao L. Cai P. Zhu H. Liu R. Wang Y. Wei Y. Huang G. Liang J. Gao X. Biomaterials. 2014;35:8854–8866. doi: 10.1016/j.biomaterials.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Chen T. Wong Y. S. Zheng W. Bai Y. Huang L. Colloids Surf., B. 2008;67:26–31. doi: 10.1016/j.colsurfb.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Luo H. Wang F. Bai Y. Chen T. Zheng W. Colloids Surf., B. 2012;94:304–308. doi: 10.1016/j.colsurfb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Abd-Rabou A. A. Shalby A. B. Ahmed H. H. Biol. Trace Elem. Res. 2019;187:80–91. doi: 10.1007/s12011-018-1360-8. [DOI] [PubMed] [Google Scholar]
- Xu C. Qiao L. Guo Y. Ma L. Cheng Y. Carbohydr. Polym. 2018;195:576–585. doi: 10.1016/j.carbpol.2018.04.110. [DOI] [PubMed] [Google Scholar]
- Nguyen T. H. D. Vardhanabhuti B. Lin M. Mustapha A. Food Control. 2017;77:17–24. doi: 10.1016/j.foodcont.2017.01.018. [DOI] [Google Scholar]
- Liu W. Li X. Wong Y. S. Zheng W. Zhang Y. Cao W. Chen T. ACS Nano. 2012;6:6578–6591. doi: 10.1021/nn202452c. [DOI] [PubMed] [Google Scholar]
- Yazdi M. H. Sepehrizadeh Z. Mahdavi M. Shahverdi A. R. Faramarzi M. A. Nano Biomed. Eng. 2016;8:246–267. [Google Scholar]
- Ranjitha V. R. Ravishankar V. R. Pharm. Nanotechnol. 2018;6:61–68. doi: 10.2174/2211738505666171113141010. [DOI] [PubMed] [Google Scholar]
- Zhong L. Zhang X. Covasa M. World J. Gastroenterol. 2014;20:7878. doi: 10.3748/wjg.v20.i24.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L. Yuan Q. Zhu H. Li Y. Guo Q. Wang Q. Bi X. Gao X. Biomaterials. 2011;32:6515–6522. doi: 10.1016/j.biomaterials.2011.05.032. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Li X. Huang Z. Zheng W. Fan C. Chen T. Nanomedicine. 2013;9:74–84. doi: 10.1016/j.nano.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Yang F. Tang Q. Zhong X. Bai Y. Chen T. Zhang Y. Li Y. Zheng W. Int. J. Nanomed. 2012;7:835–844. doi: 10.2217/nnm.12.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Zhu H. Li X. Liu Z. Zheng W. Chen T. Yu B. Wong K.-H. J. Agric. Food Chem. 2013;61:9859–9866. doi: 10.1021/jf403564s. [DOI] [PubMed] [Google Scholar]
- Pi J. Jin H. Liu R. Song B. Wu Q. Liu L. Jiang J. Yang F. Cai H. Cai J. Appl. Microbiol. Biotechnol. 2013;97:1051–1062. doi: 10.1007/s00253-012-4359-7. [DOI] [PubMed] [Google Scholar]
- Pi J. Yang F. Jin H. Huang X. Liu R. Yang P. Cai J. Bioorg. Med. Chem. Lett. 2013;23:6296–6303. doi: 10.1016/j.bmcl.2013.09.078. [DOI] [PubMed] [Google Scholar]
- Ahmed H. H. Khalil W. K. B. Hamza A. H. Toxicol. Mech. Methods. 2014;24:593–602. doi: 10.3109/15376516.2014.956912. [DOI] [PubMed] [Google Scholar]
- Resch U. Schichl Y. M. Sattler S. de Martin R. Biochem. Biophys. Res. Commun. 2008;375:156–161. doi: 10.1016/j.bbrc.2008.07.142. [DOI] [PubMed] [Google Scholar]
- Chen L. Meng Y. Guo X. Sheng X. Tai G. Zhang F. Cheng H. Zhou Y. Apoptosis. 2016;21:1291–1301. doi: 10.1007/s10495-016-1287-5. [DOI] [PubMed] [Google Scholar]
- Devarajan A. Su F. Grijalva V. Yalamanchi M. Yalamanchi A. Gao F. Trost H. Nwokedi J. Farias-Eisner G. Farias-Eisner R. Fogelman A. M. Reddy S. T. Cell Death Discovery. 2018;9:392. doi: 10.1038/s41419-018-0395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorieux C. Calderon P. B. Biol. Chem. 2017;398:1095–1108. doi: 10.1515/hsz-2017-0131. [DOI] [PubMed] [Google Scholar]
- De Moreno De LeBlanc A. LeBlanc J. G. Perdigón G. Miyoshi A. Langella P. Azevedo V. Sesma F. J. Med. Microbiol. 2008;57:100–105. doi: 10.1099/jmm.0.47403-0. [DOI] [PubMed] [Google Scholar]
- Kim T. H. Hur E. G. Kang S. J. Kim J. A. Thapa D. Mie Lee Y. Ku S. K. Jung Y. Kwak M. K. Cancer Res. 2011;71:2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- Ravagnan L. Roumier T. Kroemer G. J. Cell. Physiol. 2002;192:131–137. doi: 10.1002/jcp.10111. [DOI] [PubMed] [Google Scholar]
- Schmitt E. Parcellier A. Ghiringhelli F. Casares N. Gurbuxani S. Droin N. Hamai A. Pequignot M. Hammann A. Moutet M. Fromentin A. Kroemer G. Solary E. Garrido C. Cancer Res. 2004;64:2705–2711. doi: 10.1158/0008-5472.CAN-03-2475. [DOI] [PubMed] [Google Scholar]
- Ferreira C. A. Ni D. Rosenkrans Z. T. Cai W. Nano Res. 2018;11:4955–4984. doi: 10.1007/s12274-018-2092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkusre P. J. Nanomed. Nanotechnol. 2014;05:194. [Google Scholar]
- Gandin V. Khalkar P. Braude J. Fernandes A. P. Free Radical Biol. Med. 2018;127:80–97. doi: 10.1016/j.freeradbiomed.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Shreenath A. P., Ameer M. A. and Dooley J., Selenium Deficiency, StatPearls Publishing, 2020 [PubMed] [Google Scholar]
- Shakibaie M. Shahverdi A. R. Faramarzi M. A. Hassanzadeh G. R. Rahimi H. R. Sabzevari O. Pharm. Biol. 2013;51:58–63. doi: 10.3109/13880209.2012.710241. [DOI] [PubMed] [Google Scholar]
- Yamazaki K. Oncol. Rep. 2000;7:977–982. doi: 10.3892/or.7.5.977. [DOI] [PubMed] [Google Scholar]
- So S. S. Y. Wan M. L. Y. El-Nezami H. Curr. Opin. Oncol. 2017;29:62–72. doi: 10.1097/CCO.0000000000000342. [DOI] [PubMed] [Google Scholar]
- Yu J. Zha J. Exp. Ther. Med. 2010;1:603–610. doi: 10.3892/etm_00000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra D. P. Yeh K. Brockman R. W. Cancer Res. 1981;41:168–175. [PubMed] [Google Scholar]
- Kurek E. Ruszczyńska A. Wojciechowski M. Łuciuk A. Michalska-Kacymirow M. Motyl I. Bulska E. Rocz. Panstw. Zakl. Hig. 2016;67:253–262. [PubMed] [Google Scholar]
- Vichai V. Kirtikara K. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Tiptiri-Kourpeti A. Fitsiou E. Spyridopoulou K. Vasileiadis S. Iliopoulos C. Galanis A. Vekiari S. Pappa A. Chlichlia K. Antioxidants. 2019;8:377. doi: 10.3390/antiox8090377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyridopoulou K. Fitsiou E. Bouloukosta E. Tiptiri-Kourpeti A. Vamvakias M. Oreopoulou A. Papavassilopoulou E. Pappa A. Chlichlia K. Molecules. 2019;24:2612. doi: 10.3390/molecules24142612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyridopoulou K. Tiptiri-Kourpeti A. Lampri E. Fitsiou E. Vasileiadis S. Vamvakias M. Bardouki H. Goussia A. Malamou-Mitsi V. Panayiotidis M. I. Galanis A. Pappa A. Chlichlia K. Sci. Rep. 2017;7:3782. doi: 10.1038/s41598-017-03971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]