Figure 2.
Epitranscriptome signatures of PTSD-like fear acquisition in the amygdala
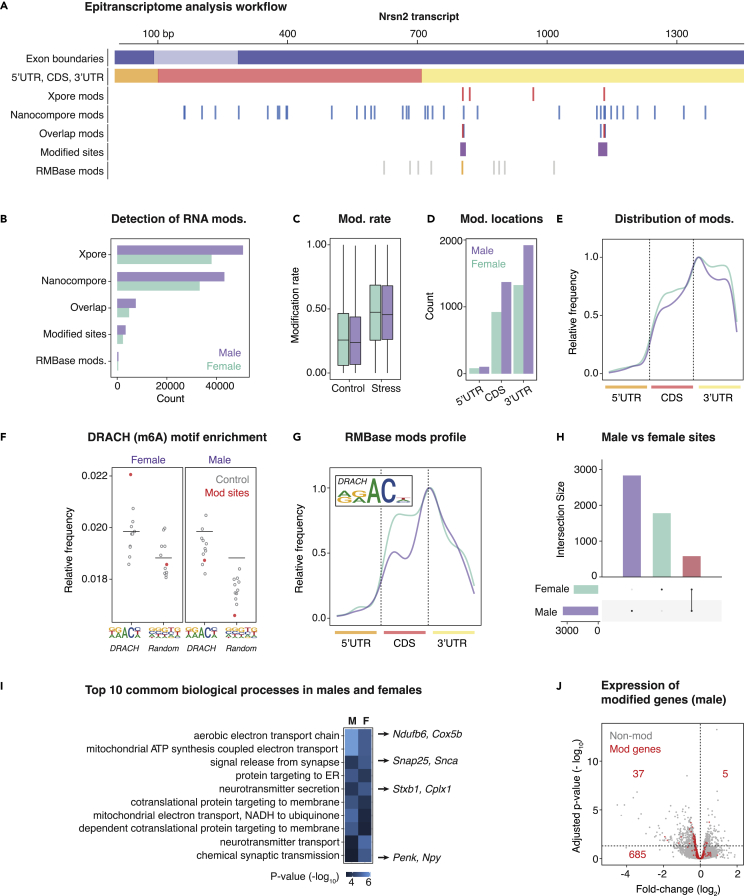
(A) Schematic example using the Nrsn2 mRNA transcript to illustrate the analytic workflow for profiling RNA modifications in conditioned versus control mice. Briefly, candidate modifications were identified separately by Xpore and Nanocompore. Only overlapping/adjacent candidates identified by both tools were retained, and multiple candidates within 10 nt were collapsed into high-confidence modified sites. Finally, modified sites were cross-checked against RMBase.
(B) The number of candidates and modified sites retained at each step of the workflow just described.
(C) The modification rates recorded by Xpore for modified sites in stress versus control and male versus female samples.
(D) The total number of modified sites identified within different regions (5′UTR, CDS and 3′UTR) of mRNA transcripts.
(E) The positional distribution of modified sites, averaged across the whole transcriptome, within the same regions (5′UTR, CDS and 3′UTR).
(F) The relative frequency of canonical DRACH motif Kmers versus random k-mers observed within modified sites (blue), as opposed to random sites in matched transcript regions.
(G) The positional distribution and k-mer frequency profile of modified sites that also overlapped RMBase modifications, with both showing characteristic features of m6A.
(H) The number of modified sites identified that were sex specific or shared between males and females.
(I) The top ten common GO terms enriched among differentially modified genes in males and females. Example genes underlying each term are highlighted.
(J) Volcano plot showing gene expression fold-changes observed between stress versus control, with genes that were identified as differentially modified highlighted in red. Minimal overlap between differentially expressed and differentially modified genes is apparent.