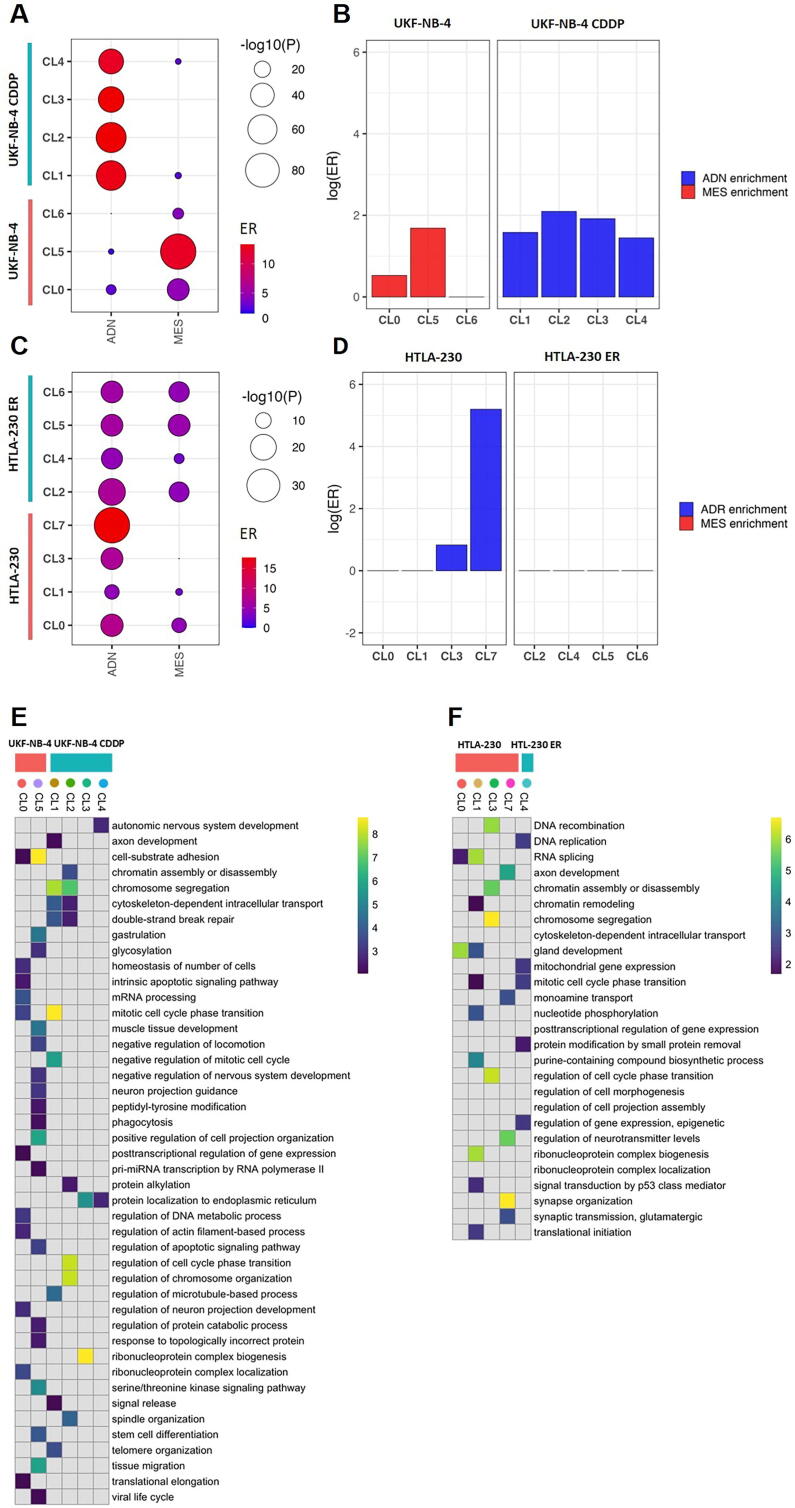
Fig. 2.
Enrichment analysis of genes expressed in the cellular clusters. (A) Balloon plot showing the enrichment of ADN and MES signatures in each cluster identified in UKF-NB-4 CDDP and UKF-NB-4, and (C) in HTLA-230 ER and HTLA-230. Balloon size indicates the statistical significance in log scale, color indicates the Enrichment Ratio (ER) from the enrichment test. (B) Plot showing Log(ER) results of comparison analysis of MES and ADN signatures expressed in the cellular clusters of UKF-NB-4 CDDP and UKF-NB-4 and (D) HTLA-230 ER and HTLA-230.(E) Enrichment analysis for the top cluster markers in relation to GO terms in UKF-NB-4 CDDP and UKF-NB-4, and (F) in HTLA-230 ER and HTLA-230. Parental and resistant cell lines are delineated by pink and blue boxes respectively and clusters are delineated by colored dots. GO gene sets overlapping with at least 10 markers in one, were included. Data represents significant results from WebGestalt analysis (FDR < 0.01), and shown in color scale ranging from 0 to the 99th data quantile (to avoid highly significant pathways dominating the heatmap). Not significant enrichments (FDR > 0.01) are reported in gray. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)