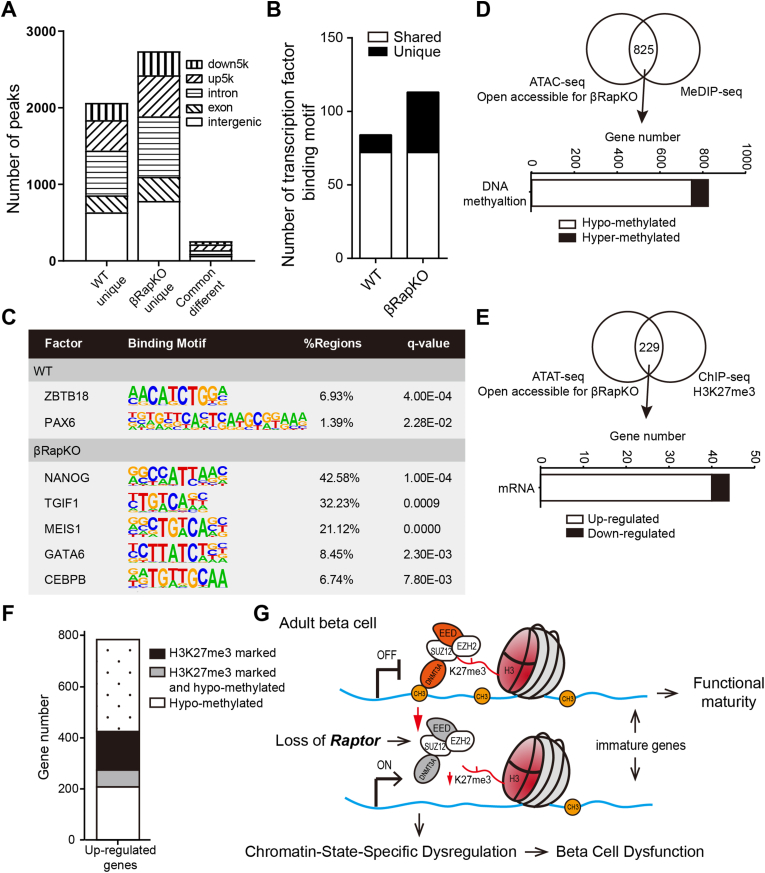
Figure 4.
Integration of ATAC-seq data with DNA methylation and H3K27me3. (A) MAnorm analysis of ATAC peaks unique open or common different in 8-week-old WT and βRapKO islet. The genomic distribution of each peak was shown with different fill pattern bars. (B) Enriched motifs of ATAC-seq peaks for transcription factor binding sites were identified by HOMER (hypergeometric test). Bar graphs of the number of transcription factor binding motifs in WT and βRapKO. White bars showed the number of transcription factor binding motifs enrich in both WT and βRapKO open chromatin regions, while black bars showed the number of unique ones. (C) Representative transcription factor binding motifs enrich in WT- or βRapKO- specific open chromatin regions. (D) 825 genes open accessible for βRapKO showed differentially DNA methylation level. Bar graph of the number of hypo- (white) and hyper-methylated (black) genes. (E) ATAC-seq analysis of WT and βRapKO showed 229 genes open accessible for βRapKO were marked with H3K27me3. H3K27me3 marked genes were identified by ChIP-seq of adult WT mice [24]. Then observing these gene expression as shown in bar graph, 40 genes showed up-regulated (white), and 4 genes down-regulated (black). (F) Bar graph showed gene number of upregulated genes in 8-week-old βRapKO mice was associated with epigenetic regulations, including DNA methylation and H3K27me3. (G) Schematic diagram of mTORC1-denpendent epigenetic regulation, including two major epigenetic mechanisms, DNA methylation and H3K27me3.