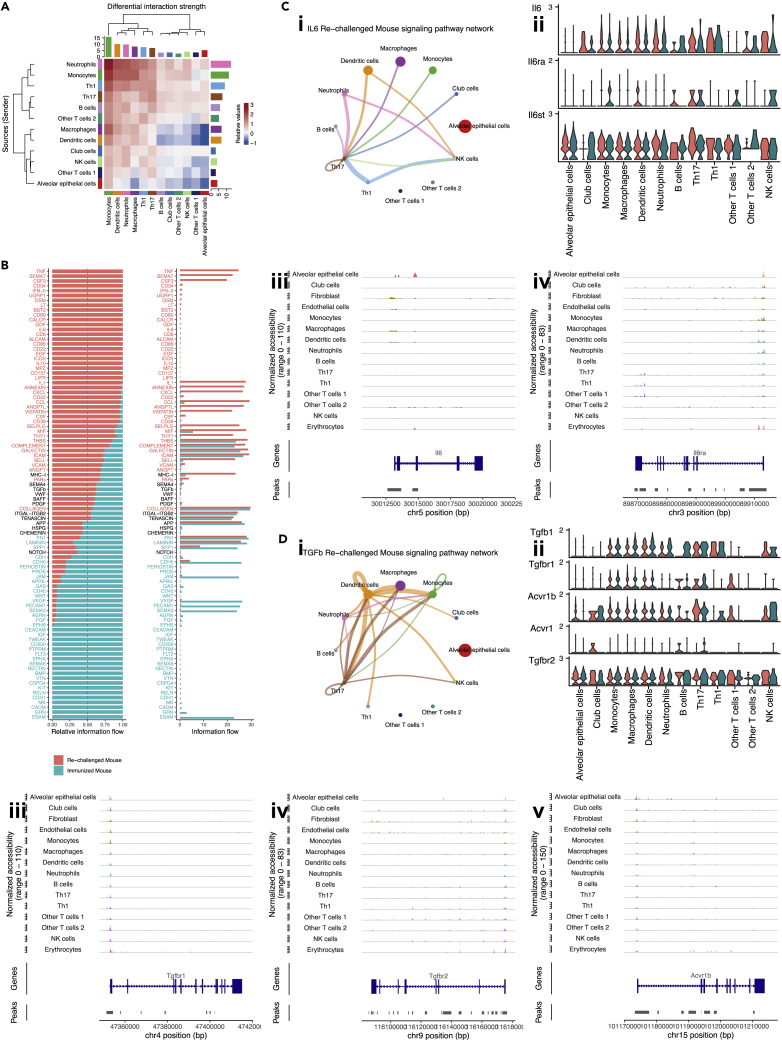
Figure 6.
Cell-cell communication among cell-type enriched spots, see also Figure S6
(A) Heatmap showing differential interaction strength between cell-type enriched spots. Outgoing signals were shown in rows, while incoming signals were shown in columns. Increased (or decreased) signals in the re-challenged mouse compared to the immunized mouse were represented using red (or blue) in the color bar. The sum of values within the same column was summarized using the colored bar plot on the top. The sum of values within the same row was summarized using the colored bar plot on the right.
(B) Bar plots showing overall information flow of each signaling pathway. Relative information flow was shown in the stacked bar plot, while raw information flow was shown in the regular bar plot. Enriched signaling pathways were colored in red or cyan.
(C) Th17 enriched spots in the re-challenged mouse were the receivers of the IL6 signaling pathway. (i) Circle plot visualizing the inferred communication network of the IL6 signaling pathway in the re-challenged mouse. The network of the IL6 signaling pathway in the immunized mouse was not significant. Circle sizes represented the number of spots in each group. Edge colors were consistent with the senders of the signal (sources), and edge weights represented the interaction strength. (ii) Violin plot visualizing the expression of genes related to IL6 signaling pathway in cell-type enriched spots from the re-challenged mouse. Gene expression from slice A3 was colored in red, and that from slice A4 was colored in cyan. (iii-iv) Peaks in genomic regions around Il6 and Il6ra in scATAC-seq data.
(D) Th17 enriched spots in the re-challenged mouse were the receivers of the TGF-β signaling pathway. (i) Circle plot visualizing the inferred communication network of the TGF-β signaling pathway in the re-challenged mouse. The network of the TGF-β signaling pathway in the immunized mouse was also significant but not shown. Circle sizes represented the number of spots in each group. Edge colors were consistent with the senders of the signal (sources), and edge weights represented the interaction strength. (ii) Violin plot visualizing the expression of genes related to TGF-β signaling pathway in cell-type enriched spots from the re-challenged mouse. Gene expression from slice A3 was colored in red, and that from slice A4 was colored in cyan. (iii-v) Peaks in genomic regions around Tgfbr1, Tgfbr2, and Acvr1b in scATAC-seq data.