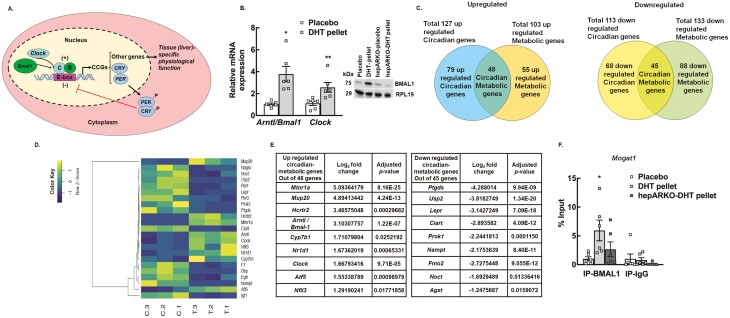
Figure 3.
High level of androgen affects the expression of a large number of hepatic circadian genes important for liver metabolism. (A) Schematic of the molecular clock. (B) Relative expression of core clock genes, Arntl1/ Bmal1 (brain and muscle arntl-like 1) and Clock (clock circadian regulator) messenger RNA levels by quantitative real-time polymerase chain reaction in livers isolated from wild-type placebo and dihydrotestosterone (DHT) pellet-treated mice. Data are displayed as means ± SE of the mean (n = 6 mice/ treatment) and normalized to Rpl19.*P ≤ 0.01 for Arntl/Bmal1, **P ≤ 0.05 for Clock vs placebo using Student’s t-test. Representative immunoblots (inset) showing total protein levels of BMAL1 in liver samples isolated from wild-type mice treated with placebo pellet, DHT pellet, and hepatocyte-specific androgen receptor (AR) knockout mice treated with placebo or DHT pellet. RPL19 protein level was used as internal control. (C) The total number of up- and downregulated metabolic genes regulated by the hepatic circadian clock was identified by comparing the total number of differentially regulated circadian and metabolic genes. (D) Hierarchical clustering is shown as a heatmap of differentially expressed circadian metabolic genes sorted by adjusted P-value by plotting their log2 transformed expression values in samples (n = 3 mice/treatment). (E) List of top 9 up- and downregulated circadian-metabolic genes. (F) Anti-BMAL1 chromatin immunoprecipitation assay in livers isolated from wild-type placebo and DHT pellet-treated mice as well as hepatocyte-specific AR knockout (hepARKO) mice treated with DHT pellet, showing BMAL1 binding on Mogat1 promoter region (−104 bp from transcription start site). Immunoglobin G (IgG) represents a nonspecific antibody. Values represent percentage input. Data are displayed as mean ± SE of the mean (n = 6 mice/treatment). *P ≤ 0.05 vs placebo using 1-way analysis of variance followed by Dunnett’s multiple comparison test.