Extended Data Fig. 10 |. GPR84 agonists stimulate uptake of antibody-opsonized cancer cells.
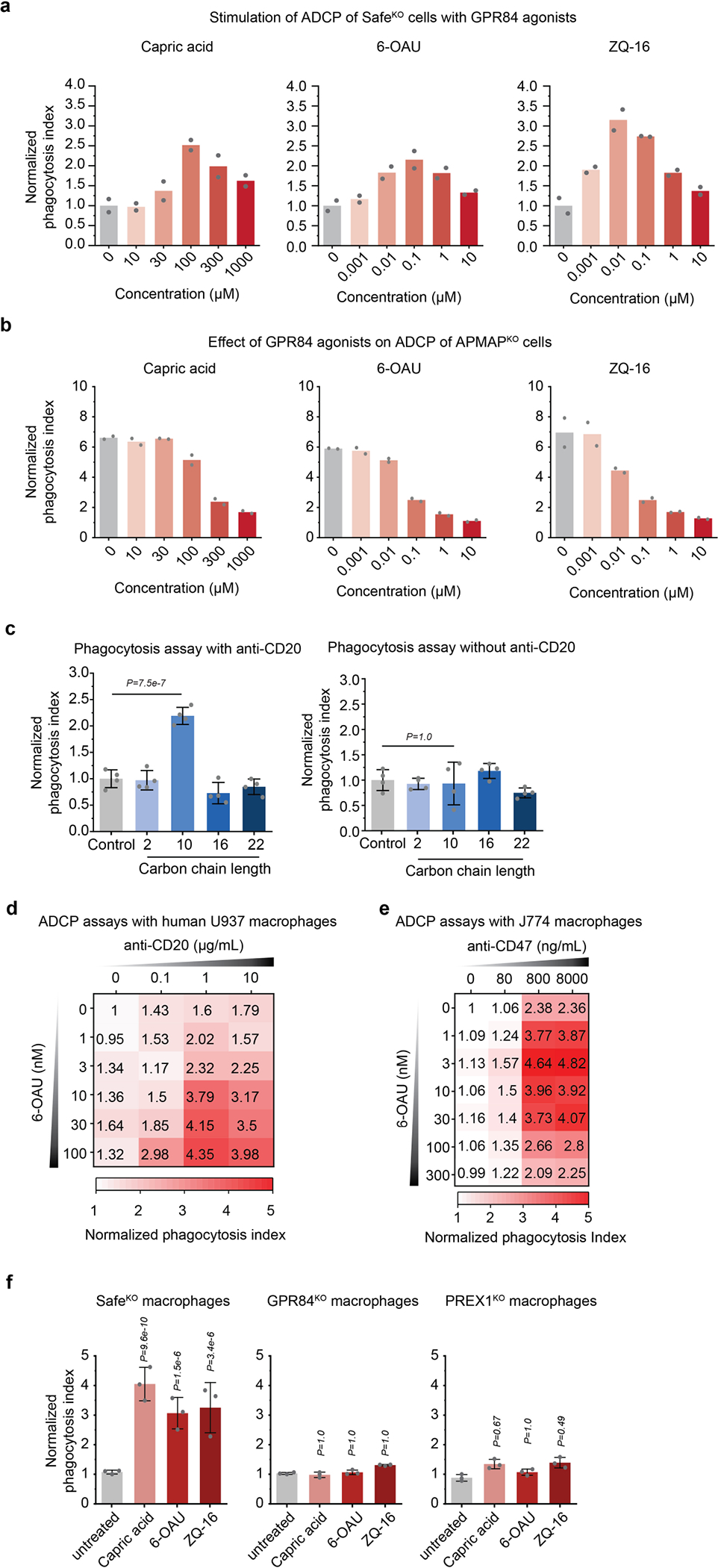
a, Phagocytosis assay for uptake of pHrodo-labelled Ramos Cas9 cells expressing Safe-targeting sgRNAs by J774 macrophages in the presence of anti-CD20 antibodies and indicated concentrations of GPR84 agonists. Data represent mean (n = 2 cell culture wells). b, Phagocytosis assay for uptake of pHrodo-labelled Ramos Cas9 cells expressing APMAP-targeting sgRNAs by J774 macrophages in the presence of anti-CD20 antibodies and indicated concentrations of GPR84 agonists. Data represent mean (n = 2 cell culture wells). c, Phagocytosis assay for uptake of pHrodo-labelled SafeKO Ramos Cas9 cells by J774 macrophages in the presence (left) or absence (right) of anti-CD20 antibodies and 100 μM saturated fatty acids of indicated carbon chain length (n = 2, acetic acid; n = 10, capric acid; n = 16, palmitic acid; n = 22, docosanoic acid). Data represent mean +/− s.d. (n = 4 cell culture wells). One-way ANOVA with Bonferroni correction. d, Heatmap of normalized phagocytosis index of Ramos cells incubated with U937 macrophages expressing indicated sgRNAs, in the presence of indicated concentrations of 6-OAU and anti-CD20. Data represent mean (n = 4 cell culture wells). e, Heatmap of normalized phagocytosis index of Ramos cells incubated with J774 macrophages expressing indicated sgRNAs, in the presence of indicated concentrations of 6-OAU and anti-CD47. Data represent mean (n = 4 cell culture wells). f, Phagocytosis assay for uptake of pHrodo-labelled Ramos Cas9 cells expressing Safe-targeting sgRNAs by J774 macrophages in the presence of anti-CD20 antibodies and GPR84 agonists (100 μM capric acid, 100 nM 6-OAU, 10 nM ZQ-16). Data represent mean +/− s.d. (n = 3 cell culture wells). One-way ANOVA with Bonferroni correction. P-values are for comparison to untreated condition for each macrophage genotype.
