Extended Data Fig. 4 |. APMAP loss sensitizes cells to ADCP in a highly specific manner and without affecting surface levels of other pro- and anti-phagocytic factors.
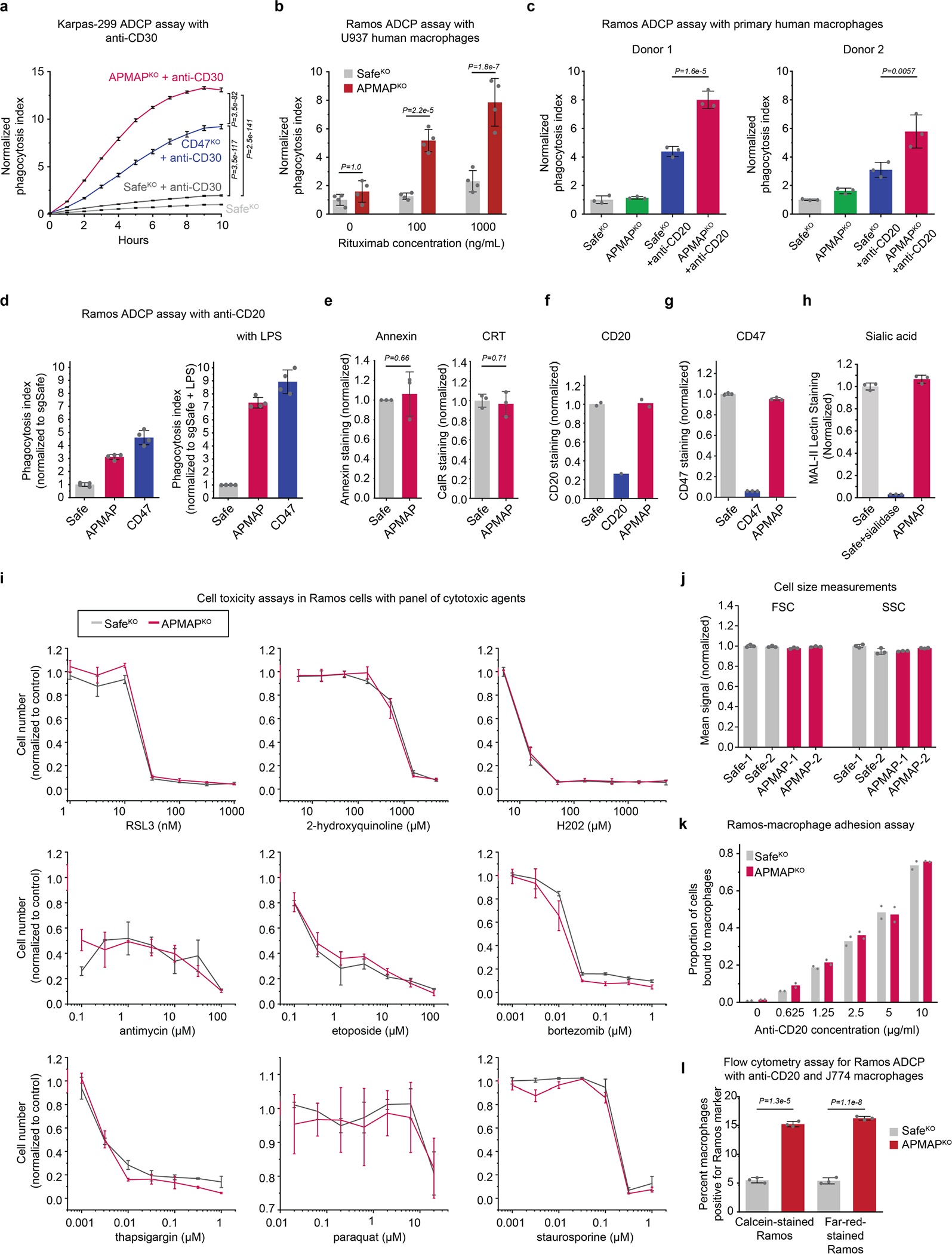
a, Phagocytosis assay for uptake of pHrodo-labelled Karpas-299 Cas9 cells expressing indicated sgRNAs by J774 macrophages in the presence or absence of anti-CD30 antibodies. Normalized phagocytosis index was calculated as average total pHrodo Red signal per well, normalized to signal in untreated control condition at final timepoint. Data represent mean +/− s.d. (n = 4 cell culture wells). Two-way ANOVA with Bonferroni correction. b, Phagocytosis assay for uptake of pHrodo-labelled Ramos cells with indicated genotypes by human U937 macrophages in the presence or absence of anti-CD20 (rituximab) antibodies at indicated concentrations. Phagocytosis index normalized to control (SafeKO) Ramos cells without anti-CD20. Data represent mean +/− s.d. (n = 4 cell culture wells). Two-way ANOVA with Bonferroni correction. c, Phagocytosis assay for uptake of pHrodo-labelled Ramos cells with indicated genotypes by human primary peripheral blood-derived macrophages, from two independent healthy de-identified human donors, in the presence or absence of 10 ng ml−1 anti-CD20 antibodies. Phagocytosis index normalized to control (SafeKO) Ramos cells without anti-CD20. Data represent mean +/− s.d. (n = 3 cell culture wells). One-way ANOVA with Bonferroni correction. d, Phagocytosis assay for uptake of pHrodo-labelled Ramos Cas9 cells expressing indicated sgRNAs by J774 macrophages with or without 24 h pre-treatment with 100 ng ml-1 LPS. Normalized phagocytosis index was calculated as average total pHrodo Red signal per well, normalized to signal in untreated control condition at final timepoint. Data represent mean +/− s.d. (n = 4 cell culture wells). e, Ramos Cas9 cells expressing indicated sgRNAs were incubated with Annexin V-FITC or anti-Calreticulin-DyLight-488 and analysed by flow cytometry. CRT, calreticulin. Data represent mean +/− s.d. (n = 3 independently stained samples). P-values were from two-tailed t-tests. f, Flow-cytometry based measurement of cell surface levels of CD20 in Ramos Cas9 cells expressing indicated sgRNAs. Data represent mean (n = 2 independently stained samples, except cells expressing CD20 sgRNA (n = 1)). g, Flow-cytometry based measurement of cell surface levels of CD47 in Ramos Cas9 cells expressing indicated sgRNAs. Data represent mean +/− s.d. (n = 3 independently stained samples). h, Flow-cytometry based measurement of cell surface levels of sialic acid in Ramos Cas9 cells expressing indicated sgRNAs. Where indicated, cells were treated with sialidase as a positive control. Data represent mean +/− s.d. (n = 3 independently stained samples). i, Viability assays (measured as cell confluence after 72 h on Incucyte, normalized to untreated SafeKO control cells) of indicated Ramos cells in the presence of indicated concentrations of 9 drugs. Data represent mean +/− s.d. (n = 3 cell culture wells). j, Flow-cytometry based measurement of forward scatter (FSC) and side scatter (SSC) in Ramos Cas9 cells expressing indicated sgRNAs. Data represent mean +/− s.d. (n = 3 independently analysed samples). k, Ramos-J774 adhesion assay in the presence of indicated antibody concentrations, using indicated GFP+ Ramos Cas9 knockout cells. Data represent mean +/− s.d. (n = 2 cell culture wells). l, Flow-cytometry based measurement of ADCP of Ramos Cas9 cells expressing indicated sgRNAs and stained with either calcein or CellTrace-Far-Red dye before incubation with J774 macrophages and anti-CD20 for 24h. Data represent mean +/− s.d. (n = 3 cell culture wells). Two-tailed t-tests were used to compare SafeKO and APMAPKO cells within each labeling condition.
