Extended Data Fig. 5 |. APMAP localizes to the endoplasmic reticulum and its cytosolic domain, transmembrane domain, and N-glycosylation are not required for its function in ADCP.
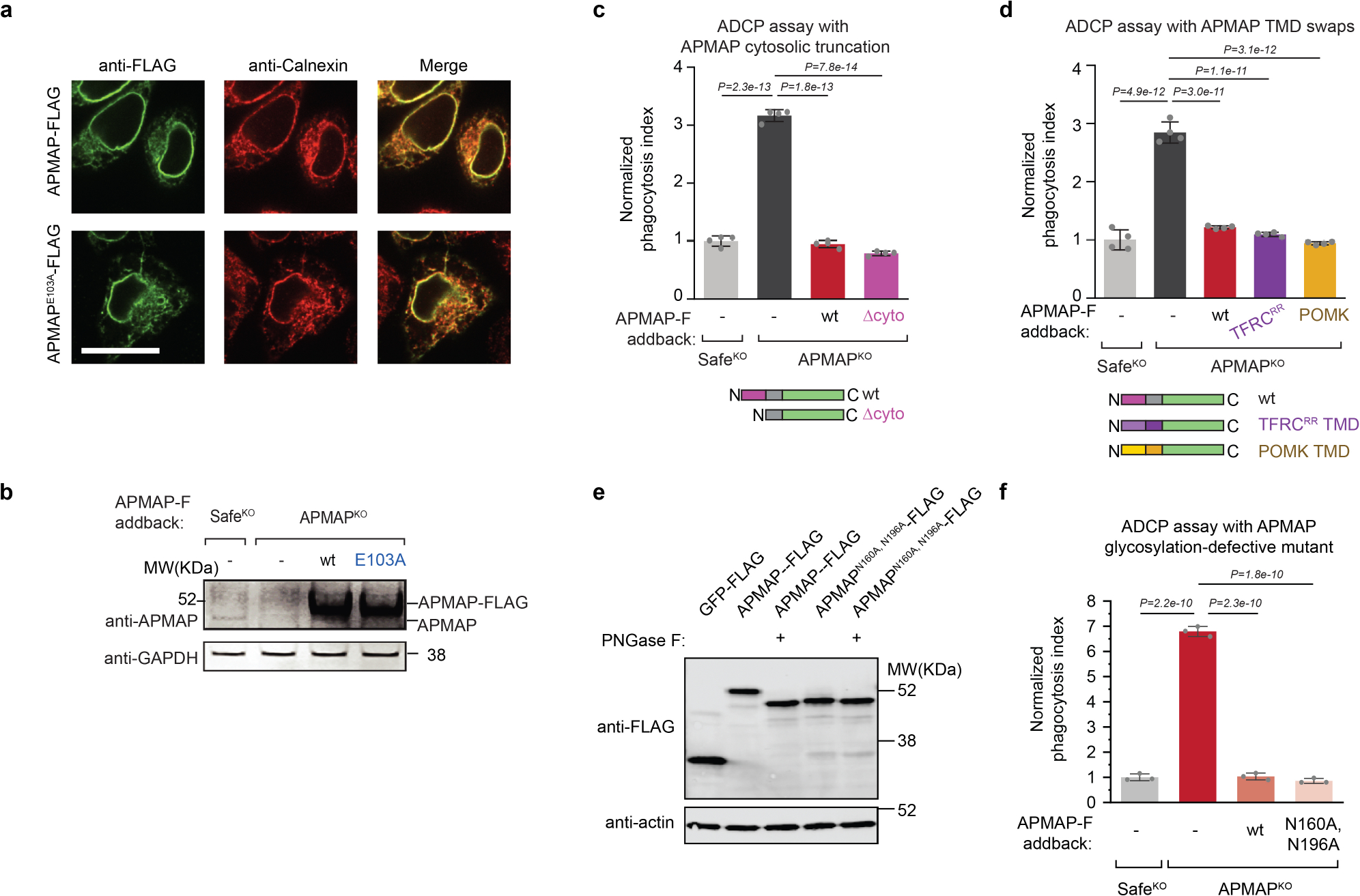
a, Localization of APMAP-FLAG and APMAPE103A-FLAG to the endoplasmic reticulum in HeLa cells. Scale bar, 20 μm. Calnexin is used as a marker of the endoplasmic reticulum. FLAG staining was representative of two independent experiments. b, Immunoblotting of cell extracts derived from Ramos cells of indicated genotypes expressing indicated APMAP-FLAG constructs. GAPDH served as loading control. Experiment was performed twice. c, d, Phagocytosis assay for uptake of pHrodo-labelled Ramos-Cas9 cells with indicated genotypes by J774 macrophages in the presence of anti-CD20 antibodies. APMAP-F, APMAP-FLAG. TFRCRR, mutant allele of TFRC that localizes primarily to the endoplasmic reticulum47. Phagocytosis index normalized to control (SafeKO) cells. Data represent mean +/− s.d. (n = 4 cell culture wells). One-way ANOVA with Bonferroni correction. e, Immunoblotting of cell extracts that were treated, where indicated, with PNGase F to remove N-glycosylation. Actin served as loading control. Experiment was performed once. f, Phagocytosis assay for uptake of pHrodo-labelled Ramos Cas9 cells expressing indicated sgRNAs and indicated addback constructs by J774 macrophages in the presence of anti-CD20 antibodies. Normalized phagocytosis index was calculated as average total pHrodo Red signal at 5 h for each well, normalized to signal in SafeKO cells at 5 h timepoint. Data represent mean +/− s.d. (n = 3 cell culture wells). One-way ANOVA with Bonferroni correction. For gel source data, see Supplementary Figure 1.
