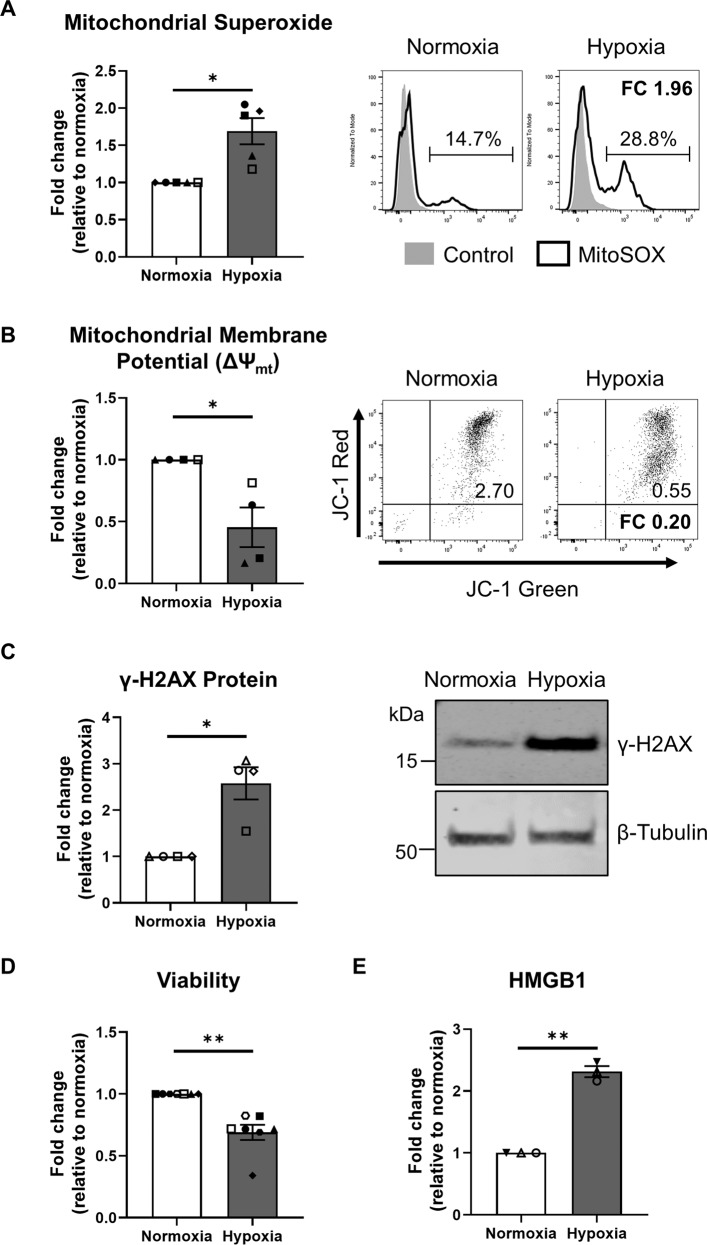
Fig. 1. Hypoxia induces pathways of mitochondrial oxidative damage/dysfunction and DNA damage in human primary PTEC.
A Left panel: fold changes (relative to normoxia) in mitochondrial superoxide levels (% MitoSOX+ cells) for PTEC cultured under normoxic and hypoxic conditions. Bar graphs represent mean ± standard error mean (SEM). Symbols represent individual donor PTEC; n = 5. *P < 0.05, paired t test. Right panel: representative MitoSOX staining (black unfilled) compared with unstained control (grey filled) for PTEC cultured under normoxic and hypoxic conditions. Mitochondrial superoxide levels (% MitoSOX+ cells) are presented for each histogram, with fold change (FC) value relative to normoxic PTEC also shown. B Left panel: Fold changes (relative to normoxia) in mitochondrial membrane potential [measured as ratio of ΔMFI JC-1 red/ΔMFI JC-1 green; with delta median fluorescence intensity (ΔMFI) representing MFI test – MFI unstained control] for PTEC cultured under normoxic and hypoxic conditions. Bar graphs represent mean ± SEM. Symbols represent individual donor PTEC; n = 4. *P < 0.05, paired t test. Right panel: representative JC-1 dot plots of PTEC cultured under normoxic and hypoxic conditions. Mitochondrial membrane potential (ΔΨmt) values are presented for each histogram, with fold change (FC) value relative to normoxic PTEC also shown. C Left panel: fold changes (relative to normoxia) in γ-H2AX protein levels (as a ratio of loading control β-tubulin) for PTEC cultured under normoxic and hypoxic conditions. Bar graphs represent mean ± SEM. Symbols represent individual donor PTEC; n = 4. *P < 0.05, paired t test. Right panel: western blot for γ-H2AX for PTEC cultured under normoxic and hypoxic conditions (15 µg total protein per lane). Representative images from one of four donor PTEC are presented; full and uncropped western blot available as Supplementary Material. D, E Fold changes (relative to normoxia) in cell viability (MTT assay) (D) and HMGB1 (ELISA) (E) for PTEC cultured under normoxic and hypoxic conditions. Bar graphs represent mean ± SEM. Symbols represent individual donor PTEC; n = 7 for viability data and n = 3 for HMGB1 data. **P < 0.01, paired t test.