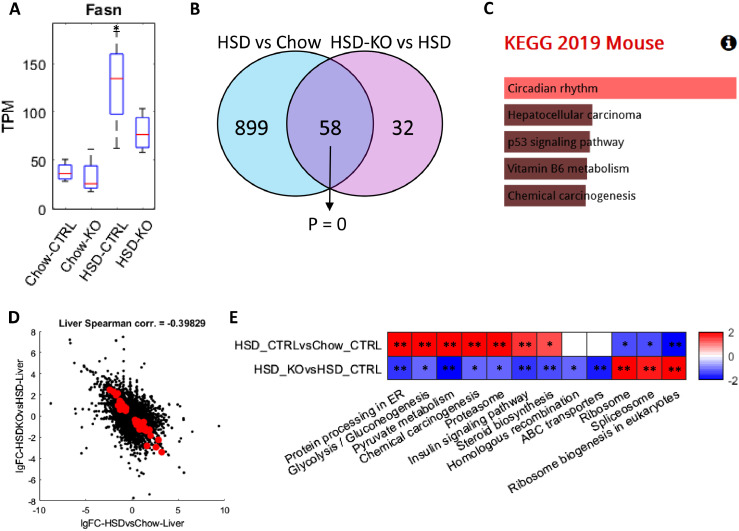
Figure 2.
Analysis for liver transcriptomics profile of 8-week wild-type and Pkl KO mice fed with CHOW and HSD. (A) Box plot showing the expression of Fasn in different conditions, and the asterisks indicated the significant increase of Fasn expression compared to wild-type mice fed with CHOW (P < 0.05). (B) Venn-diagram showing the number and hypergeometric P value of overlapped DEGs between HSD Ctrl vs CHOW Ctrl and HSD Ctrl vs HSD KO in liver. (C) Barplot showing the significantly enriched KEGG pathways of the overlapped DEGs in Figure 2B. (D) Scatter plot showing the correlation between the log 2 fold changes of gene expression of all genes in HSD Ctrl vs CHOW Ctrl (x-axis) and HSD KO vs HSD Ctrl (y-axis), where the dots highlighted in red are the overlapped DEGs highlighted in Figure 2B. (E) Heatmap showing the differentially expressed pathways in the two comparisons, respectively, where each cell in the plot shows the minus log 10 P value if they are up-regulated, and positive log 10 P value if they are down-regulated (* P adj. <0.1; ** P adj. <0.05). Only pathways that are significantly differentially expressed in HSD KO vs HSD Ctrl (P adj. <0.1) are displayed and pathways without significant expression alterations (P adj. >0.1) are marked white.