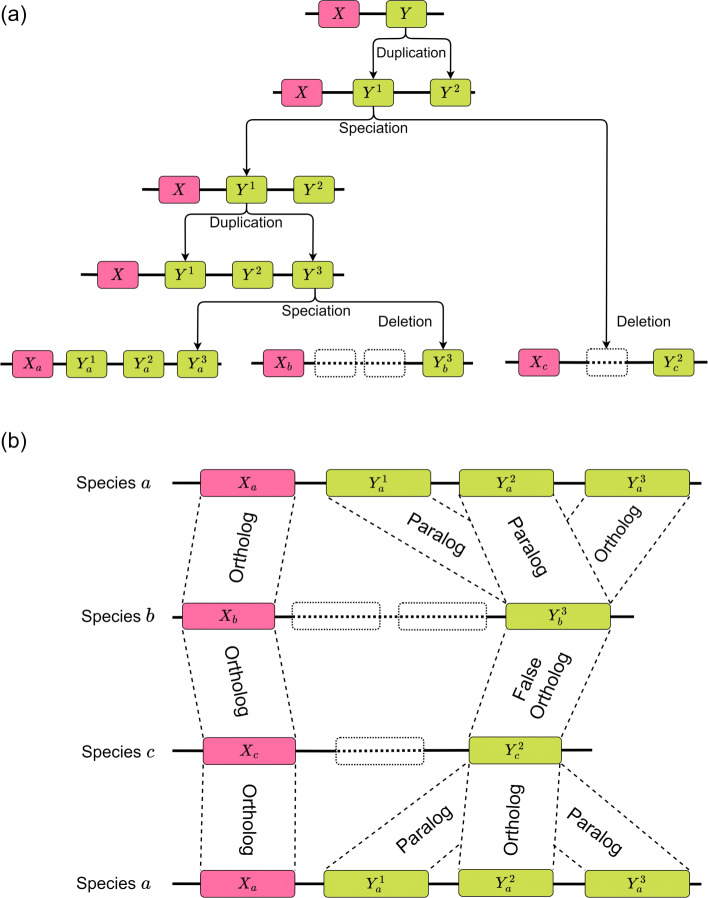
Fig. 1.
An example of evolution resulting in different classes of homology. In (a), deletion and speciation events are depicted, resulting in both orthologs and paralogs. The resulting relationships between all pairs of X segments as well as between all pairs of Y segments are depicted in (b). Notably, segments and participate in three important homology relationship types. As the most recent common ancestor of and is at a speciation event, they are orthologous to each other. Segments and on the other hand are paralogs, as their homology is a result of duplication. Finally, and are special types of paralogs known as “false orthologs” i.e. paralogous segments all of whose other copies in their respective genomes have since been deleted, resulting in two segments which appear to never have been duplicated. The presence of such false orthologies complicates the problem of core-genome alignment particularly, but also the problem of further categorizing homologies identified through genome alignment. It is worth noting that homology relationships within a single species’ genome do exist, but are not depicted here. For example, and are all paralogous to each other