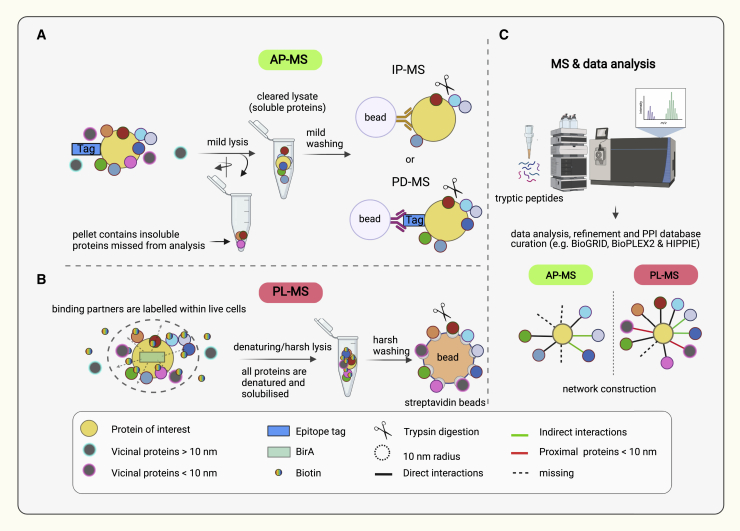
Figure 2.
Schematic of AP-MS and PL-MS approaches
(A) AP-MS using an IP-grade antibody (IP-MS) or epitope tag (PD-MS). Epitope-tagged proteins of interest (yellow circle) are endogenously or ectopically expressed in target cells. After mild lysis, solubilized proteins are separated from insoluble proteins by centrifugation. Soluble proteins are mixed with beads conjugated to specific antibodies against the epitope tag or POI to capture the associated proteins. After several stringent washes, direct and indirect interactions (colored circles) are co-purified with the bait from the complex cell lysate.
(B) In PL-MS, BirA-fused proteins of interest (yellow circle and light green rectangle) are endogenously or ectopically expressed in target cells. Labeling is initiated with the addition of biotin to cultured cells. BirA enzyme mediates the labeling of direct and indirect interactions as well as vicinal proteins within 10 nm distance (represented by dotted circle). After labeling, cells are lysed using a harsh and denaturing lysis buffer to enhance solubilization of proteins. Biotinylated proteins are immobilized on streptavidin beads and then are washed before proceeding to on-bead tryptic digestion.
(C) Peptides generated in (A) and (B) are desalted using C18 columns and then subjected to MS analysis. As depicted in the interaction network, PL-MS results in increased detection of interactions and fewer missed interactions than AP-MS.