As a result of the work of many laboratories, a new paradigm describing the manner by which bacteria respond to repair DNA damage has emerged. This paradigm holds that under any growth condition, essentially all replication forks formed at oriC encounter DNA damage and either stall or collapse before they can complete synthesis of the genome. Maintenance of cell viability therefore requires both correction of the DNA lesion via the action of the DNA repair enzymes and replication fork restart via the combined action of the DNA recombination and replication enzymes. A proposal has been advanced to distinguish this pathway, which operates as a housekeeping function in the absence of exogenous insult to the cell and is likely to be inherently nonmutagenic, from the SOS response, which is induced by exogenous DNA damage and includes error-prone repair, that it be named CPR for coordinated processing of damaged replication forks (M. M. Cox, M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians, submitted for publication). In this minireview, we will describe the central role of PriA in the replication fork restart step of CPR.
THE BEGINNINGS
PriA was discovered because of its requirement for the synthesis of the complementary strand of bacteriophage φX174 single-stranded DNA in vitro (42, 49). This reaction could be divided into two steps: synthesis of a RNA primer and elongation of that primer to make the complementary strand. The latter step was catalyzed by the replicase of Escherichia coli, the DNA polymerase III holoenzyme. The former step required a number of proteins, now referred to as the φX-type primosomal proteins. These were ultimately resolved into PriA, PriB, PriC, DnaT, DnaB, DnaC, and DnaG (26, 27).
The priming step required the assembly of a replication intermediate on the DNA that was capable of both movement along the single strand and synthesis of the primer. This protein machine was named the primosome (2). Assembly of the primosome on DNA was specific in that only bacteriophage DNAs that used the φX-type primosomal proteins for replication could support assembly of the primosome. This specificity resided in PriA, which recognized and bound to a site on φX DNA to serve as a scaffold for assembly of the primosome (7, 42). This site was named a primosome assembly site (PAS) (27) and shown to be a region of the DNA that could assume a stable hairpin structure (42).
Intensive biochemical studies revealed many interesting facets of primosome assembly and function. DnaB and DnaG were demonstrated to be the cellular replication fork DNA helicase (3, 17, 32) and Okazaki fragment primase (4, 48), respectively. DnaG interacts distributively with the preprimosome (the primosome without DnaG) through a specific interaction with DnaB (45, 51). PriA also has 3′→5′ DNA helicase activity (16, 20), which is the opposite directionality of the DnaB helicase activity (17). Thus, the preprimosome is capable of both bidirectional translocation along single-stranded DNA and bidirectional DNA helicase activity (21).
The primosome concept—an enzyme or a group of enzymes that exist as a complex and that can both unwind duplex DNA and catalyze the synthesis of short oligoribonucleotide primers—was an attractive way to provide two of the necessary activities at a replication fork. Indeed, the φX-type primosome was shown to be able to provide just those activities at replication forks during both rolling-circle DNA replication, using specialized tailed form II DNA templates (32, 51), and the replication of ColE1-type plasmid DNAs (31).
However, the relationships of PriA, PriB, PriC, and DnaT to cellular DNA replication were unclear. Whereas the primosome activities would seem to be necessary for DNA replication, none of these proteins were required for oriC-directed replication of small plasmid templates reconstituted with purified proteins (10) and none of the genes encoding these proteins have been identified in exhaustive searches for DNA replication mutants with a slow stop phenotype. These seemingly contradictory data can be reconciled by recent studies showing that whereas these proteins are unlikely to be present in a primosome formed at oriC through the action of DnaA, they are crucial for completing chromosomal DNA replication. In fact, the combination of PriA, PriB, PriC, and DnaT appears to play a role similar to that of DnaA in that they load DnaB into the forming replisome. Whereas DnaA does this at oriC, as discussed below, PriA, PriB, PriC, and DnaT do this at recombination intermediates.
GENETICS AND MODELS
The gene encoding PriA was molecularly cloned by reverse genetics (19, 35). The open reading frame specified a protein of 732 amino acids having a calculated molecular mass of 81.7 kDa. PriA has the seven amino acid motifs common to most DNA helicases and falls into the SF2 superfamily, where it defines its own subgroup (6). Between helicase motifs IV and V is an extensive Cys metal-binding motif. This is an unusual insertion in a DNA helicase and presumably is responsible for some of the unique properties of the protein.
Several interesting priA missense mutant genes have been isolated and/or constructed by site-directed mutagenesis on plasmids so that the mutant proteins could be overproduced, purified, and studied in vitro. In some cases, these alleles have been transferred to the chromosome (S. J. Sandler, P. Nurse, J. Liu, and K. J. Marians, unpublished data). Some conclusions from these studies include the following. (i) PriA is a multifunctional protein with both ATPase, helicase, and translocase activities. These are genetically separate from its primosome assembly activity (54). (ii) Only the primosome assembly function is apparently uniquely needed for the role of PriA in the cell (14, 39, 54). (iii) The cysteine-rich region thought to be involved in Zn2+ binding is important for helicase activity (55) and interactions with at least one other primosome assembly protein, PriB (23).
In order to assess the role of PriA in chromosomal DNA replication, disruptions of priA were constructed (18, 37). priA1::kan has the kanamycin resistance-encoding gene inserted downstream of codon 405 accompanied with a deletion of codons 406 to 466 (18). priA2::kan has the gene encoding kanamycin resistance inserted between codons 154 and 155, 70 amino acids upstream of the nucleotide-binding motif (37). The strains with both of these disruptions are devoid of PriA replication activity as measured biochemically.
The priA disruption strains had severely reduced viability and filamented extensively (18, 37). Lee and Kornberg (18) noted that priA1::kan strains were UV sensitive. We found that priA2::kan strains were constitutively induced for the SOS response and that some viability could be restored if a sulA mutation was also present (37). Complete suppression of SOS induction could be achieved if the priA300 allele was provided in trans (54). This mutant gene encodes PriA K230R, a PriA protein that is no longer a DNA helicase but can still assemble a primosome (54). To account for the observed induction of SOS in the absence of exogenous DNA-damaging agents, we proposed that PriA primosome assembly activity was required to restart replication forks that had stalled because of encountering endogenous DNA damage (37, 53). The mechanism of replication fork assembly emerged as a result of subsequent investigation of the phenotypes of priA mutants.
priA strains have a very complex phenotype. In addition to those listed above, they are sensitive to rich media (28) and are defective in homologous recombination (14, 39), both inducible and constitutive stable DNA replication (iSDR and cSDR, respectively) (28), and in the repair of both double-strand breaks (14) and UV-damaged DNA (14, 18, 39). As was the case for the induction of the SOS response, all of these phenotypes could be suppressed by providing priA300 in trans. As discussed below, it is likely that all of these phenotypes arise either as a result of a failure to assemble replication forks at recombination intermediates or because of the accumulation of recombination intermediates in the cell.
priA null mutants also acquire suppressor mutations rapidly that restore viability, recombination proficiency, and UV and X-ray resistance to wild-type levels (14, 39). Seventeen independent suppressor mutations have all been mapped to dnaC (39). DnaC is another primosomal protein that forms a complex with DnaB in solution (50) and facilitates the delivery of DnaB to single-stranded DNA.
How to explain all of these phenotypes based on the loss of the PriA primosome assembly activity? A common thread among many of these phenotypes is that they involved recombination. Sensitivity to rich media presumably results from the added complexities of having multiple chromosomes in the same cell, providing a rich milieu for recombination. Models of repair of double strand breaks and some modes of repair of UV-damaged DNA (43) involve the use of sister chromosomes to recover the lost genetic information via the establishment of recombination intermediates. Both iSDR and cSDR require recA. iSDR also requires other recombination genes such as recB, recC, and recF, whereas cSDR does not require any other recombination genes (13). This engendered proposals that PriA was directing the assembly of the φX-type primosome at recombination intermediates which, in turn, led to the establishment of a replication fork (14, 39).
The defect in homologous recombination during such processes as P1 transduction could be explained in one of two ways. Either replication fork assembly at recombination intermediates is an obligatory step for resolution (i.e., “ends-out” RecBCD-mediated recombination [44]) or the presence of recombination intermediates themselves block replication fork progression, creating a requirement for restart.
Masai et al. proposed that the initiating structures during iSDR and cSDR were a D loop and a R loop, respectively, targeted by PriA (28). A genetic interaction between recG and priA also suggested that D loops were the target for PriA (1). RecG is required for wild-type levels of recombination and repair (25). This protein is a 3′→5′ DNA helicase that recognizes Holliday junctions and three-strand junctions such as those found in D loops (47). RecG can catalyze branch migration in vitro so as to drive the invading strand in a D loop in the 3′→5′ direction into the donor duplex, thereby helping to establish Holliday intermediates (46).
Suppressors of the recombination and repair defects of recG mutations, named srgA, were found to be allelic with priA (1). Because priA+ overexpressed from a multicopy plasmid had a dominant-negative effect on srgA suppression of recG mutant phenotypes and overexpression of priA300 did not, Al-Deib et al. (1) concluded that the necessary event for creation of a suppressor was a reduction in the helicase activity of PriA. These researchers envisioned that at a Holliday structure, the two enzymes could catalyze branch migration in opposite directions. When RecG was present, the net result was positive for DNA repair. When the helicase activity of RecG was absent but that of PriA was present, the net result was counterproductive for DNA repair. Whether PriA has this function in recG+ cells remains to be determined. Nevertheless, the affinity of PriA for a D loop has been demonstrated biochemically and is central to its ability to direct replication fork restart.
PRIA-DIRECTED REPLICATION FORK ASSEMBLY AT D LOOPS
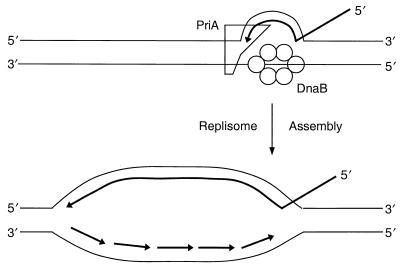
McGlynn et al. (29) first showed that PriA could specifically bind a D loop composed of short oligonucleotides, whereas it did not bind the corresponding bubble structure. Our subsequent analyses indicated that PriA binding of the D loop was by virtue of its affinity for bent DNA at three-strand junctions (36). The affinity of the protein for junctions with a 5′ tail was very high, with an equilibrium dissociation constant (KD) of 1.3 nM, 30-fold greater than the affinity for a junction with a 3′ tail. In a D loop with a 3′ invading strand, this places PriA at the junction formed by the 3′ end of the invading strand and the target DNA (Fig. 1). Preferential binding of PriA to three-strand junctions was also noted by Jones and Nakai (9).
FIG. 1.
PriA-directed replisome assembly at a D loop. (Top) Relative positions of PriA and DnaB on D-loop DNA during assembly of the primosome, as deduced from DNA footprinting studies. (Bottom) Direction of subsequent replication fork progression. The invading strand is used as the primer for leading-strand synthesis, whereas Okazaki fragment synthesis is primed as a result of the periodic interaction of DnaG with DnaB as the helicase moves 5′→3′ along the lagging-strand template.
For replication fork assembly to occur on a D loop, the 3′-OH of the invading strand can be used as the leading-strand primer. The direction of replication fork propagation then requires that the functional primosome be bound to and moving on the displaced strand (Fig. 1). DNA footprinting studies showed that PriA actually bound to all three strands of the D loop at the three-strand junction (22). Although a footprint of a complete primosome on D-loop DNA could not be obtained, alterations in nuclease sensitivity indicating that unwinding of the D loop had occurred confirmed that PriA could direct the assembly of a primosome on this structure (22). Unwinding of the D loop was consistent with DnaB being loaded onto the displaced strand. The disposition of the other primosomal proteins in the complex remains unclear. Jones and Nakai (9) have also demonstrated φX-type primosome-dependent unwinding of a three-strand junction during phage Mu replicative transposition.
Using a circular, nicked, double-stranded template carrying a D loop, we demonstrated that PriA could direct replication fork assembly at this structure (24). Replication fork formation required the action of all the primosomal proteins in the presence of the single-stranded DNA-binding protein (SSB), although the dependence on PriC was weak (see below for a possible interpretation of this). That a bona fide replisome had formed was supported by the observation that rapid replication fork propagation required the protein-protein interaction between DnaB and the τ subunit of the DNA polymerase III holoenzyme. This interaction has been shown both to define the replisome, directing which one of the two polymerase cores in the holoenzyme becomes the leading-strand polymerase (11, 52), and to stimulate the unwinding rate of DnaB (12, 52).
Replication fork assembly proceeded as well in the presence of the PriA K230R protein as in the presence of the wild-type PriA protein (24). Because provision of priA300 in trans suppresses all of the priA2::kan phenotypes, this suggests that the ability of PriA to direct replication fork assembly at a recombination intermediate like a D loop is likely to represent its primary role in the cell. This is also supported by the properties of the DnaC810 protein (L. Xu and K. J. Marians, submitted for publication). This protein is encoded by dnaC810, a naturally arising suppressor of all of the phenotypes of the priA2::kan allele (39).
DnaC810 was able to bypass the action of PriA, PriB, PriC, and DnaT and assemble a replication fork on the D-loop template in the presence of only DnaB and the holoenzyme (24). Because the presence of DnaC in a DnaB-DnaC complex prevents DnaB binding to single-stranded DNA and transfer of DnaB to SSB-coated DNA cannot occur, access of DnaB to the DNA in the cell is effectively limited to targeting by mechanisms that involve additional DNA replication proteins to generate a SSB-free region of single-stranded DNA for DnaB binding. Two mechanisms that operate in vivo are known to do this: DnaA-directed initiation of DNA replication at oriC (10) and PriA-directed assembly of a primosome at recombination intermediates such as a D loop as described above. The underlying gain-of-function of the mutant DnaC810 is an ability to transfer DnaB from a DnaB-DnaC complex directly to SSB-coated DNA (Xu and Marians, submitted).
A key feature of DnaC810-mediated bypass of PriA function that remains to be illuminated is whether this protein has also assumed the targeting role provided by PriA in bringing DnaB to the D loop. For complete primosome assembly, the specificity of PriA for binding to the D loop assures proper targeting. Does DnaC810 have a similar specificity, or is another, as yet unknown protein(s) needed?
The importance to the cell of replication fork assembly at recombination intermediates is underscored by the biochemical properties of the DnaC810 protein and by recent data indicating the existence of multiple parallel pathways for replication fork restart.
MULTIPLE PATHWAYS OF REPLICATION FORK RESTART
Assembly of the φX-type primosome on φX174 viral DNA is an ordered process. As analyzed by gel mobility shift and enhanced chemiluminescence-Western analyses using a 304-nucleotide single-stranded DNA carrying the PAS from φX DNA as the substrate, PriA-directed primosome assembly proceeded in discrete steps resulting in, sequentially, the following protein-DNA complexes (33): (i) PriA-PAS DNA, (ii) PriA-PriB-PAS DNA, (iii) PriA-PriB-DnaT-PAS DNA, (iv) PriA-PriB-DnaT-DnaB-PAS DNA (the preprimosome), and (v) PriA-PriB-DnaT-DnaB-DnaG-DNA (the primosome). The association of DnaG with the preprimosome is transient and is governed by a protein-protein interaction between DnaG and DnaB (45). Although PriC was not required for stable preprimosome assembly on the 304-nucleotide PAS DNA substrate, it was found as a component of preprimosomes assembled on and isolated bound to full-length φX viral DNA (34). Thus, it is possible that PriC plays a role in stabilizing the primosome on DNA, rather than being required in an absolute fashion for primosome assembly. Is this the pathway of primosome assembly at recombination intermediates in the cell? Probably not.
The pathway of assembly of the φX-type primosome outlined above predicted that mutations in priB and priC would display phenotypes identical to those of priA2::kan. This was not the case. Neither the ΔpriB302 nor priC303::kan mutations exhibit any of the phenotypes of the priA2::kan mutation, although the priB mutant does shows a twofold decrease in homologous recombination (38). Remarkably, however, when the mutations were combined, the priB priC double mutant was barely viable and grew even more slowly than strains carrying the priA2::kan mutation. Like priA2::kan strains, the priB priC strains also acquired suppressor mutations rapidly.
Partial phenotypic rescue occurs when dnaC809 (resulting in the same amino acid substitution as dnaC810) is combined with ΔpriB302 and priC303::kan. These triple-mutant strains were SOS induced and recombination defective similar to priA2::kan strains but were UVr and as viable as the wild type. A second suppressor mutation was selected in the triple-mutant background that restored the strain to essentially wild-type properties. This suppressor was found to also arise in dnaC (the doubly mutated gene is called dnaC809,820). The dnaC820 mutation maps just two codons downstream of the dnaC809 mutation. These results suggested that priC and priB encoded redundant functions in parallel pathways (Fig. 2). It is very likely that the selective activity of dnaC809 and dnaC809,820 reflects the fact that they are compensating for the absence of different, redundant pathways of replication fork assembly at recombination intermediates.
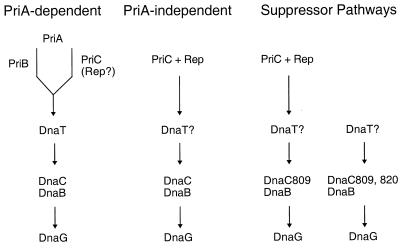
FIG. 2.
Multiple pathways of replication fork assembly at recombination intermediates. One possible set of pathways, based on the genetic data discussed in the text, involves the replication restart primosomal proteins and Rep during replication fork assembly at recombination intermediates. Each column denotes one possible pathway. The steps separated by vertical arrows in each column represent the order of action of particular proteins during assembly of the restart primosome. This order of assembly is based on biochemical data discussed in the text. The bifurcation in the PriA-dependent pathway indicates that PriA can interact with either PriB or PriC (and Rep?). Question marks indicate that the action of this protein in the pathway is unclear. Regardless of which proteins are present on the recombination intermediate as a final protein-DNA complex, the object of each pathway is to place DnaB in position on single-stranded DNA so that a new replication fork can form.
A strong case can be made that PriA is the major force in directing replication fork restart in E. coli. However, why, then, is priA2::kan not lethal to the cell? One possibility is that there are multiple pathways of replication fork restart. As suggested by the data described above, different pathways may utilize different assortments of the φX-type primosomal proteins. In their ongoing studies of mechanisms that lead to the generation of double-strand breaks in vivo, Seigneur et al. (41) demonstrated that rep recB combinations were lethal and suggested that the absence of Rep led to more-frequent replication fork arrest. This group had shown previously that fork arrest led to the production of double-strand breaks (30). At this level, however, replication fork arrest cannot be distinguished from the failure to restart, so it was not a total surprise that Seigneur et al. (41) also reported that the rep priA combination was also lethal.
Like PriA, Rep is a 3′→5′ DNA helicase. Interestingly, rep was identified as a gene that, when mutated, did not support the growth of φX174 (5), and biochemical analysis showed that it could form a replication fork with the φX gene A protein and the holoenzyme that was responsible for viral strand production during the phage life cycle (40). Replication forks in rep mutants have been reported to travel at one-half the speed of the wild type (15). This is consistent with a role for Rep in replication fork restart.
Using an indirect selection technique, we have searched for synthetic lethality between PriA and other primosomal proteins. We confirmed that priA rep formed a lethal combination and further demonstrated that the priA priC combination was also lethal, whereas both the priA priB and the priC rep combinations were viable (S. J. Sandler, unpublished data). Moreover, the dnaC809,820 mutation, but not the dnaC809 mutation, could suppress either synthetically lethal combination. Taken together, this suggests that there are both PriA-dependent and -independent pathways of replication fork restart at recombination intermediates. The PriA-dependent pathway requires at least PriA and either PriB or PriC, whereas the PriA-independent pathway requires at least both PriC and Rep. One model for how these pathways might function is given in Fig. 2. This model may also explain why the requirement for PriC during primosome assembly on φX DNA in vitro is weak.
PERSPECTIVES
It is now clear that replication fork restart is an essential cellular function. Given that the role of the φX174-type primosome is now reasonably well established within the process of fork restart, it seems appropriate to suggest that it be identified by a name that is more descriptive. We therefore propose to call the primosome that requires PriA, PriB, PriC, DnaT, DnaB, DnaC, and DnaG for assembly the replication restart primosome. Even so, the complete picture detailing replication fork reactivation must still be elucidated. What are the other players in the PriA-dependent and -independent pathways? This should become clear from an approach combining biochemical and genetic methods. Clearly in vitro systems are now required that reconstitute, e.g., double-strand break repair. Further genetic dissection of the restart pathways may be aided by screens for suppressors of priA point mutations that we have recently transferred to the genome and which recapitulate the phenotypes of priA2::kan (Sandler et al., unpublished).
Are PriA and Rep functional equivalents? If they are, is there a role for the Rep DNA helicase activity during fork restart? At the moment, this seems unlikely because the priA300 Δrep::kan combination is viable (Sandler, unpublished). It would be interesting to see whether this pair of mutations could be combined with recG mutations. Interestingly, unlike the case with replication fork restart, PriA DNA helicase activity is required for growth of phage Mu (8). It has been suggested that the PriA helicase first operates to create a single-stranded region on the Mu recombination intermediate to which DnaB can then bind (9).
How does the type of recombination intermediate and/or the assortment of recombination proteins present on the intermediate influence the choice of restart pathway taken? These are murky waters. There are currently clear differences between the biochemical phenotypes of the DnaC suppressor proteins and the phenotypes of their respective alleles. DnaC810 cleanly bypasses PriA, PriB, PriC, and DnaT function in replication fork assembly on the D-loop template (24) and can deliver DnaB to SSB-coated DNA without the aid of other proteins (Xu and Marians, submitted), yet the genetic observations suggest a PriC dependence (38; Sandler, unpublished data). And in vitro, DnaC810,820 shows a PriC dependence in replication reactions containing a SSB-coated template, DnaB, DnaG, and the holoenzyme (L. Xu and K. J. Marians, unpublished data), whereas the genetics would argue against this requirement (38; Sandler, unpublished; Sandler et al., unpublished). It seems likely that this apparent disparity relates to the fact that our biochemical systems are not yet complex enough to mirror the situation in vivo with complete accuracy.
ACKNOWLEDGMENTS
Studies from our laboratories were supported by start-up funds from the University of Massachusetts and grant RPG-99-194-01-GMC from the American Cancer Society (S.J.S.) and NIH grant GM34557 (K.J.M.).
REFERENCES
- 1.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicase of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai K-I, Kornberg A. Unique primed start of phage φX174 replication and mobility of the primosome in a direction opposite chain synthesis. Proc Natl Acad Sci USA. 1981;78:69–73. doi: 10.1073/pnas.78.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T A, Sekimizu K, Funnell B E, Kornberg A. Helicase action of DnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987;262:6877–6885. [PubMed] [Google Scholar]
- 4.Bouché J-P, Zechel K, Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J Biol Chem. 1975;250:5995–6001. [PubMed] [Google Scholar]
- 5.Denhardt D T, Dressler D H, Hathaway A. The abortive replication of φX174 DNA in a recombination-deficient mutant of Escherichia coli. Proc Natl Acad Sci USA. 1967;57:813–817. doi: 10.1073/pnas.57.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationship. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 7.Greenbaum J, Marians K J. The interaction of Escherichia coli replication factor Y with complementary strand origins of DNA replication: contact points revealed by DNase footprinting and protection from methylation. J Biol Chem. 1984;259:2594–2601. [PubMed] [Google Scholar]
- 8.Jones J M, Nakai H. The φX174-type primosome promotes replisome assembly at the site of recombination in bacteriophage Mu transposition. EMBO J. 1997;16:6886–6895. doi: 10.1093/emboj/16.22.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones J M, Nakai H. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J Mol Biol. 1999;289:503–515. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- 10.Kaguni J M, Kornberg A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell. 1984;38:183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Dallmann H G, McHenry C S, Marians K J. τ protects β in the leading-strand polymerase complex at the replication fork. J Biol Chem. 1996;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Dallmann H G, McHenry C S, Marians K J. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 13.Kogoma T. Stable DNA replication. Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kogoma T, Cadwell G W, Barnard K G, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane H E D, Denhardt D T. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol. 1974;97:99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 16.Lasken R S, Kornberg A. The primosomal protein n′ of Escherichia coli is a DNA helicase. J Biol Chem. 1988;263:5512–5518. [PubMed] [Google Scholar]
- 17.LeBowitz J H, McMacken R. The Escherichia coli dnaB protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 18.Lee E H, Kornberg A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee E H, Masai H, Allen G C, Jr, Kornberg A. The priA gene encoding the primosomal replicative n′ protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M S, Marians K J. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc Natl Acad Sci USA. 1987;84:8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M S, Marians K J. The Escherichia coli primosome can translocate actively in either direction along a DNA strand. J Biol Chem. 1989;264:14531–14542. [PubMed] [Google Scholar]
- 22.Liu J, Marians K J. PriA-directed assembly of a primosome on D loop DNA. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Nurse P, Marians K J. The ordered assembly of the φX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271:15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Xu L, Sandler S J, Marians K J. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd R G, Buckman C. Genetic analyses of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marians K J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 27.Marians K J. Enzymology of DNA replication in prokaryotes. Crit Rev Biochem. 1984;17:153–215. doi: 10.3109/10409238409113604. [DOI] [PubMed] [Google Scholar]
- 28.Masai H, Asai T, Kubota Y, Arai K-I, Kogoma T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 30.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minden J S, Marians K J. Replication of pBR322 DNA in vitro with purified proteins: requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985;260:9316–9325. [PubMed] [Google Scholar]
- 32.Mok M, Marians K J. The Escherichia coli preprimosome and DnaB helicase can form replication forks that move at the same rate. J Biol Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 33.Ng J Y, Marians K J. The ordered assembly of the φX174-type primosome. I. Isolation and identification of intermediate protein-DNA complexes. J Biol Chem. 1996;271:15642–15648. doi: 10.1074/jbc.271.26.15642. [DOI] [PubMed] [Google Scholar]
- 34.Ng J Y, Marians K J. The ordered assembly of the φX174-type primosome. II. Preservation of primosome composition from assembly through replication. J Biol Chem. 1996;271:15649–15655. doi: 10.1074/jbc.271.26.15649. [DOI] [PubMed] [Google Scholar]
- 35.Nurse P, DiGate R, Zavitz K H, Marians K J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci USA. 1990;87:4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurse P, Liu J, Marians K J. Two modes of PriA binding to DNA. J Biol Chem. 1999;274:25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 37.Nurse P, Zavitz K H, Marians K J. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandler S J, Marians K J, Zavitz K H, Coutu J, Parent M A, Clark A J. dnaC mutations suppress defects in DNA replication and recombination associated functions in priB and priC double mutants in E. coli K-12. Mol Microbiol. 1999;34:91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- 39.Sandler S J, Sawra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott J F, Eisenberg S, Bertsch L L, Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage φX174: catalytic strand separation in advance of replication. Proc Natl Acad Sci USA. 1977;74:193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seigneur M, Bidnenko V, Ehrlich S E, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 42.Shlomai J M, Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in φX174 DNA. Proc Natl Acad Sci USA. 1980;77:799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith G R. Homologous recombination in prokaryotes. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith G R. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell. 1989;58:807–809. doi: 10.1016/0092-8674(89)90929-x. [DOI] [PubMed] [Google Scholar]
- 45.Tougu K, Peng H, Marians K J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 46.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitby M C, Vincent S, Lloyd R G. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickner S. DNA or RNA priming of bacteriophage G4 synthesis by Escherichia coli dnaG protein. Proc Natl Acad Sci USA. 1977;74:2815–2819. doi: 10.1073/pnas.74.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickner S, Hurwitz J. Association of φX174 DNA-dependent ATPase activity with an E. coli protein, replication factor Y, required for in vitro synthesis of φX174 DNA. Proc Natl Acad Sci USA. 1975;72:3342–3346. doi: 10.1073/pnas.72.9.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickner S, Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci USA. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu C A, Zechner E L, Marians K J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- 52.Yuzhakov A, Turner J, O'Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 53.Zavitz K H, Marians K J. Dissecting the functional role of PriA protein-catalyzed primosome assembly in Escherichia coli DNA replication. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 54.Zavitz K H, Marians K J. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]
- 55.Zavitz K H, Marians K J. Helicase-deficient cysteine to glycine substitution mutants of Escherichia coli replication protein PriA retain single-stranded DNA-dependent ATPase activity. Zn2+ stimulation of mutant PriA helicase and primosome assembly activities. J Biol Chem. 1993;268:4337–4346. [PubMed] [Google Scholar]