Abstract
The COVID-19 pandemic has been fueled by SARS-CoV-2 novel variants of concern (VOC) that have increased transmissibility, receptor binding affinity, and other properties that enhance disease. The goal of this study is to characterize unique pathogenesis of the Delta VOC strain in the K18-hACE2-mouse challenge model. Challenge studies suggested that the lethal dose of Delta was higher than Alpha or Beta strains. To characterize the differences in the Delta strain’s pathogenesis, a time-course experiment was performed to evaluate the overall host response to Alpha or Delta variant challenge. qRT-PCR analysis of Alpha- or Delta-challenged mice revealed no significant difference between viral RNA burden in the lung, nasal wash or brain. However, histopathological analysis revealed high lung tissue inflammation and cell infiltration following Delta- but not Alpha-challenge at day 6. Additionally, pro-inflammatory cytokines were highest at day 6 in Delta-challenged mice suggesting enhanced pneumonia. Total RNA-sequencing analysis of lungs comparing challenged to no challenge mice revealed that Alpha-challenged mice have more total genes differentially activated. Conversely, Delta-challenged mice have a higher magnitude of differential gene expression. Delta-challenged mice have increased interferon-dependent gene expression and IFN-γ production compared to Alpha. Analysis of TCR clonotypes suggested that Delta challenged mice have increased T-cell infiltration compared to Alpha challenged. Our data suggest that Delta has evolved to engage interferon responses in a manner that may enhance pathogenesis. The in vivo and in silico observations of this study underscore the need to conduct experiments with VOC strains to best model COVID-19 when evaluating therapeutics and vaccines.
Introduction
The COVID-19 pandemic is being perpetuated by the emergence of “Variants of Concern” (VOC): mutant strains of SARS-COV-2 with enhanced disease-causing abilities. Early in the pandemic, strains obtained the D614G mutation that enhanced binding of the spike protein to the host hACE2 receptor. The mutation quickly became dominant among circulating strains around the world. Several potential variant strains were noted and monitored but few demonstrated any unique or enhanced differences in virulence or transmission. In September 2020, the B.1.1.7 strain was first identified in the United Kingdom and quickly spread to become the dominant variant worldwide in early 2021 [1]. Later, in December 2020, two other variants, Beta (B.1.351) and Gamma (P.1) were identified in South Africa and Japan/Brazil, respectively [2–4]. Both Beta and Gamma variants increased concerns because they decreased the efficacy of several vaccines in clinical trials [5–10].
In addition to the effects on vaccine efficacy in clinical trials, there were also reports that Alpha and Beta variants were able to cause enhanced disease in humans [11–16]. Animal studies with transgenic K18-hACE2-mice and hamsters supported those observations [17, 18]. Before circulating VOC established their dominance, two doses of Pfizer-BioNTech’s mRNA vaccine, BNT162b2, exhibited 95% efficacy in protecting individuals from severe COVID-19 in retrospective cohort studies of COVID-19-associated hospitalizations in early 2021 [19]. However, the efficacy of the BNT162b2 vaccine was slightly diminished in the context of Alpha (93.7% efficacy) [20].
As Alpha, Beta, and Gamma continued to spread, the Delta variant (B.1.617.2) was identified in India, preluding a massive surge of COVID-19 cases in the country [21]. Unsurprisingly, Delta was able to cause breakthrough cases of infection within a portion of fully vaccinated people, decreasing the overall efficacy of ChAdOx1, mRNA-1273, or BNT162b2 vaccines [20, 22–24]. In March 2021, the Delta variant was detected in the United States, and reports of its increased transmissibility necessitated investigation of its threat to both unvaccinated and vaccinated populations. Key pathogenic mutations in the B.1.617 SARS-CoV-2 lineage, which includes the Delta (B.1.617.2) and Kappa (B.1.617.1) VOC, occur within the spike protein, which mediates viral attachment and entry into host cells via the ACE2 receptor [25, 26]. These mutations have been reported to increase transmissibility among the population and decrease antibody neutralization [27, 28]. Delta does not harbor the N501Y substitution in the spike protein that was characteristic of the Alpha and Beta lineages but Delta does harbor the D614G substitution in spike, which contributes to the increased fitness and transmissibility of many VOC strains [29, 30]. Some strains of Delta are reported to harbor the K417N mutation previously found in Beta and Gamma, which sparked debate over designating the strain as a new variant [31, 32]. This mutant strain was labeled as “Delta Plus.” Additional mutations within the spike protein of Delta include P681R, which diverge from other VOC and help to enhance ACE2 receptor binding and cellular entry, contributing more to Delta’s pathogenicity [33].
To characterize the pathogenicity of Delta (utilizing strain B.1.617.2) and how it diverges from prior VOC (WA-1, Alpha, and Beta strains), we performed a time-course challenge study in K18-hACE2 transgenic mice which were intranasally challenged with Alpha and Delta strains. Through analysis of viral RNA load, tissue pathology, cytokine profiling, and total transcriptomics of the lung, we discovered indications that Delta causes increased interferon type I and II responses corresponding with greater lung inflammation when compared to Alpha. These data help to advance pre-clinical models of COVID-19 as well as serve as a benchmark to compare past and future VOC strains.
Results
The Delta variant of SARS-CoV-2 has a greater LD100 than Alpha and Beta VOCs in K18-hACE2 mice
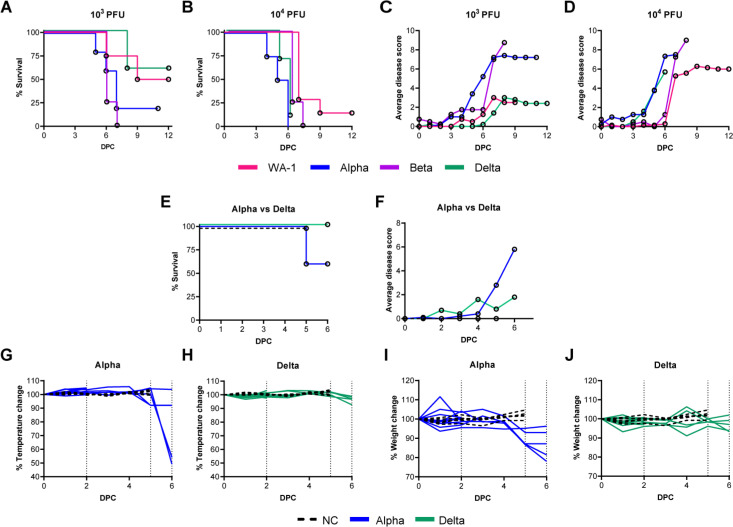
To date, most K18-hACE2-mouse or Golden Syrian hamster studies have utilized ancestral viral strains of SARS-CoV-2 (e.g., WA-1, D614G, B.1 Wuhan), and few studies have been performed with emergent VOC [2, 34–38]. Early studies in K18-hACE-mice with a variety of viral strains used a wide range of challenge doses from as low as 100 PFU to as high as 105 PFU [39–41]. In pilot studies with the Alpha and Beta variants, we evaluated 105 PFU (high dose) and observed mortality as early as day 4 post challenge, suggesting and confirming that VOCs have enhanced virulence in K18-hACE-mice compared to WA-1 ancestral strain [42]. We then aimed to identify an appropriate challenge dose for WA-1, Alpha, Beta, and Delta VOC that could cause symptomatic disease for appropriate comparisons between the subtle aspects of variant pathogenicity that can be masked by lethal challenge doses. At the low dose of 103 PFU, WA-1 challenge only resulted in 50% mortality of the mice (Fig 1A). We observed that a challenge dose of 103 PFU using Alpha or Beta resulted in 80% and 100% mortality, respectively by day 7 post challenge. Although this comparison between Alpha and Beta challenge at 103 PFU is not statistically significant (P = 0.6452), there is a significant difference between the survival of WA-1 challenged mice compared to Beta at the 103 PFU dose, where WA-1 elicits higher survival (P = 0.0446). Despite the observations of enhanced virulence of Delta in humans, 103 PFU challenge of K18-hACE2-mice mirrored WA-1 survival more so than Alpha or Beta (Fig 1A). As expected, increasing the challenge dose to 104 PFU decreased survival of K18-hACE2-mice for all strains (Fig 1B). At the 104 PFU challenge dose, Delta resulted in 0% survival (Delta vs WA-1 P = 0.0002) (Fig 1A and 1B). The 104 PFU challenge dose also shortened the time to mortality for Alpha (WA-1 vs Alpha P = 0.0008). As we suspected from an ancestral strain with reduced virulence compared to emerging variants of concern, total mortality of WA-1 infected groups was not observed at either dose. Using a disease scoring system established previously [42, 43] we observed higher average disease scores for Alpha and Beta than Delta in K18-hACE2-mice intranasally challenged with 103 PFU (Alpha vs Delta P = 0.0355; Beta vs Delta P = 0.0039) (Fig 1C and 1D). Interestingly, Delta’s observable disease phenotypes remained low by day 6 when Alpha- and Beta- challenged mice started to develop greater observable disease phenotypes consistent with morbidity (Alpha vs Delta P = 0.0056). Delta’s disease phenotypes were scored higher following 104 PFU challenge, where they progressed in a pattern that was similar to Alpha. Delta-challenged mice received higher average disease scores than Beta by day 6 post challenge (Fig 1D). These experiments suggested that Delta compared to Alpha, and Beta causes mild disease in the K18-hACE2-mouse model (at 103 PFU dose) despite its ability to cause severe disease in humans. We reasoned that these differences warranted further investigation.
Fig 1. Survival, disease scores, weight, and temperature of K19-hACE2-mice challenged with WA-1, Alpha, Beta, and Delta strains.
K18-hACE2 transgenic mice were intranasally challenged with 103 or 104 PFU WA-1, Alpha (B.1.1.7), Beta (B.1.351) or Delta (B.1.617) SARS-CoV-2, or mock-challenged with PBS (NC). Post-challenge, mouse survival (A, B) (n = 4 for 103 WA-1, n = 7 for 104 WA-1, n = 3 for 103 Alpha, n = 4 for 104 Alpha, n = 4 for 103 Beta, n = 4 for 104 Beta, n = 5 for 103 Delta and n = 10 for 104 Delta) and average disease scores per group on each day (C, D) were evaluated. Based on experimental results, mice challenged with Alpha (n = 10 split between timepoints) and Delta (n = 10 split between timepoints) or mock-challenged with PBS (n = 5), were evaluated at day 2 and day 6 post-challenge to measure survival (E), average disease scores (F), changes in rectal temperature (G, H) and bodyweight(I, J). Dotted lines indicate euthanasia points on day 2, day 5 (euthanasia of mock-challenge controls) and day 6. NC = no challenge.
Mice challenged with of Alpha or Delta experience unique disease manifestation
To gain additional insights into the differences in pathogenesis of Alpha and Delta in K18 mice, we utilized a challenge dose of 103 PFU which allows for differential disease manifestation between Alpha and Delta. Rectal temperature and weight loss were monitored in mice challenged with 103 PFU of Alpha or Delta. In agreement with their lack of mortality and low disease scores (Fig 1E and 1F), Alpha-challenged mice had significant loss of body temperature (hypothermia) and weight loss between days 5 and 6 post challenge (Fig 1G and 1H). However, Delta-challenged mice maintained body temperature and did not experience weight loss (Fig 1I and 1J). At day 6 post-challenge, Alpha challenged mice had reached morbidity based on disease scoring and required euthanasia as a humane endpoint. However, at day 6 Delta challenged mice remained below euthanasia criteria. Collectively, these data demonstrate that Alpha and Delta cause distinct disease profiles in K18-hACE2-mice.
Mice challenged with Delta have similar levels of viral RNA burden in the nares, and lungs but lower amounts of viral RNA in the brain
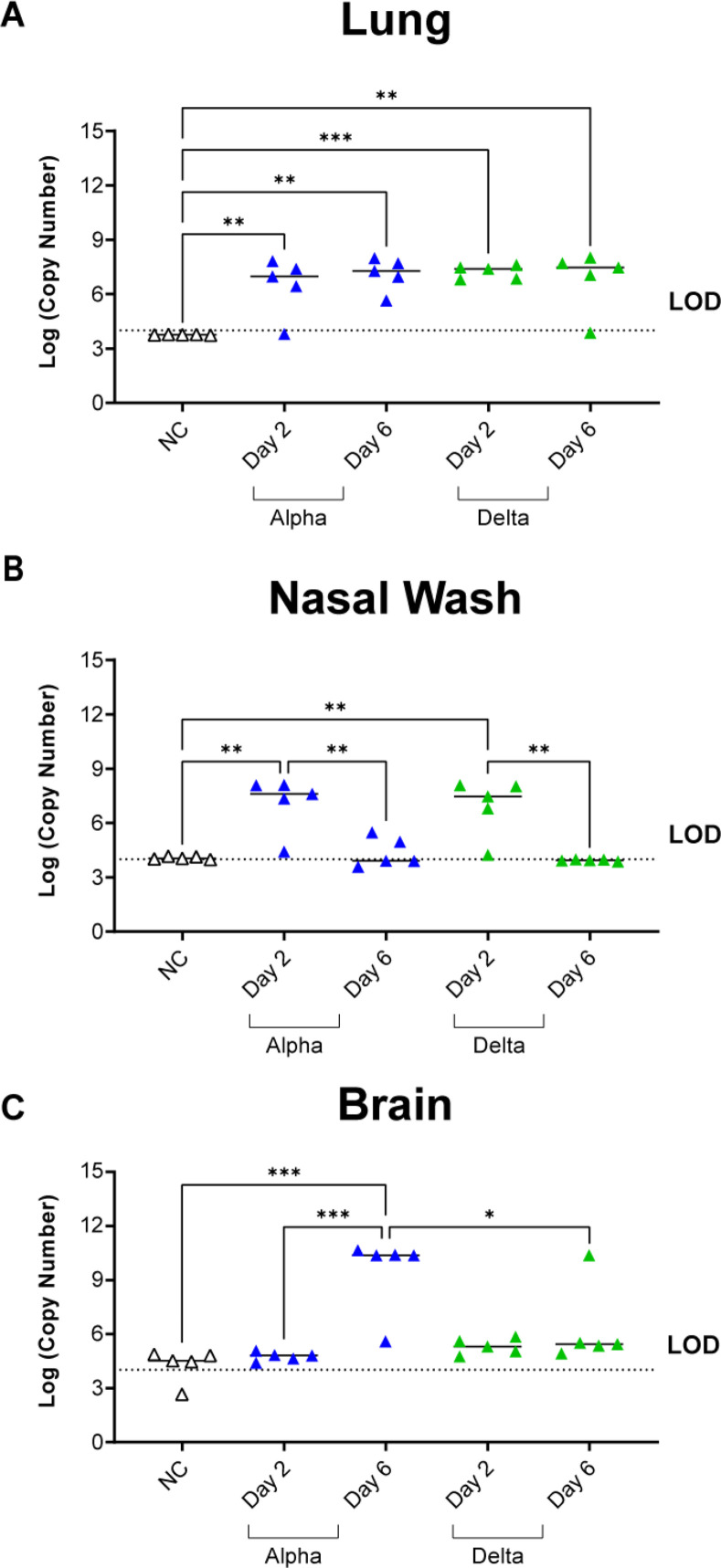
It has been previously reported that SARS-CoV-2 viral burden is highest two days after challenge in mice [44], and based on our data we recognize that the Alpha variant will cause mortality at day 6 post-challenge; however, the Delta variant challenge did not cause the same disease, or survival (Fig 1A and 1B). Therefore, we aimed to evaluate viral RNA burden at day 2 and 6 post challenge for Alpha- or Delta-challenged mice. At euthanasia on day 2 and 6 the lung, brain, and nasal wash fluid were collected from challenged mice to quantify viral RNA burden via nucleocapsid qRT-PCR. We observed that viral RNA burden was remarkably similar (not statistically significant) between Alpha and Delta in the lung at both time points (Fig 2A). In the nasal wash, viral load was also detectable at similar levels (not statistically significant) for both Alpha and Delta at day 2. In contrast to the lung, the viral RNA burden decreased to undetectable levels at day 6 for both VOC (Fig 2B). Surprisingly, challenge with Delta only resulted in one mouse with detectable viral RNA in the brain at day 6 (Fig 2C). In contrast, mice infected with Alpha exhibited low RNA levels at day 2 and a 2-fold increase at day 6, suggesting an increase in viral replication in the brain over time (Fig 2C). These observations suggest that Delta at a challenge dose of 103 PFU replicates efficiently in the airway in a manner similar to Alpha but Delta appears to have lower brain localization previously observed in K18-hACE2-mice [41, 44–46].
Fig 2. Viral RNA burden of disease-associated tissues was quantified using nucleocapsid qPCR.
Viral RNA was detectable in the lung tissue of challenged mice from both variants at both timepoints compared to no challenge (One-Way ANOVA, left to right: P = 0.0082, 0.0011, 0.0008, 0.0029) (A). In Nasal Wash, viral RNA burden increased from both variants at Day 2 compared to no challenge (One-Way ANOVA, P = 0.0016 Alpha; P = 0.0030 Delta) and was decreased at Day 6 for both variants from copy numbers at Day 2 (One-Way ANOVA, P = 0.0043 Alpha; P = 0.0019 Delta) with no statistical difference between variants at each time point (B). Alpha-challenged mice have increased detectable viral RNA in the brain at Day 6 compared to Day 2 (One-Way ANOVA, P = 0.0005) while Delta challenge results in no viral RNA burden in the brain (One-Way ANOVA: P = 0.0002 NC vs Alpha Day 6, P = 0.0228 Alpha Day 6 vs Delta Day 6) (C). Dashed lines indicate limit of detection (LOD) via qPCR. NC = no challenge. Data is representative of n = 5 mice per experimental group.
Mice challenged with Delta experience significant inflammation in the lung tissue
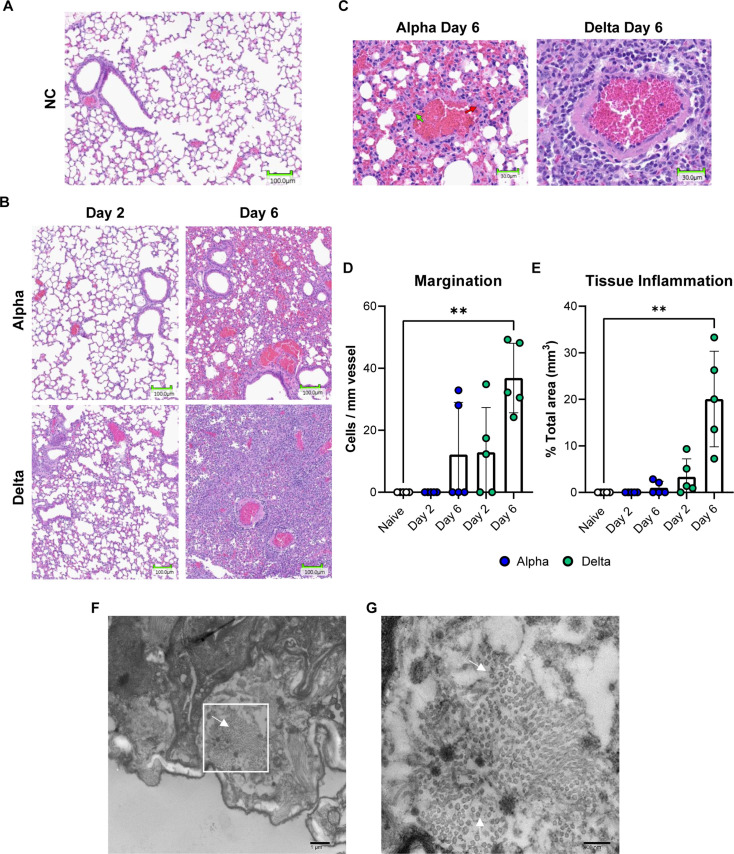
Based on qRT-PCR data showing viral RNA burden in the lung but not brain of 103 PFU Delta-challenged mice, we hypothesized that relative to Alpha, Delta may cause more robust pneumonia and less encephalitis in K18-hACE2-mice. To test this, we performed histopathological analysis on the lungs collected at days 2 and 6 to characterize the pneumonia caused by Alpha or Delta challenge. At two days following challenge, inflammation, and recruitment of inflammatory cells could already be observed in the Delta- but not Alpha-challenged mice when compared to the lungs of no challenge mice (Fig 3A and 3B). At this time, a pathologist identified that the inflammatory infiltrate was composed predominantly of lymphocytes and occasional histiocytes. By day 6, an increase in marked perivascular inflammation and margination was measured in Delta- (36.86 marginating lymphocytes per mm length of endothelium) and Alpha-challenged mice (12.19 marginating lymphocytes per mm length of endothelium) (unpaired t-test P = 0.026) (Fig 3C and 3D). In challenged Alpha lungs, less total vessels were identifiable within areas of inflammation, awarding these samples a margination score of zero cells/mm of vessel. Overall, less inflammation occurred in Alpha lungs. At day 6, only 1% of the lung area featured inflammation in Alpha, compared to 20% for Delta-challenged mice (unpaired t-test P = 0.003) (Fig 3E). Within these tissue areas (the airway, alveoli, and thin mucous layer), the presence of Delta virions was identified using electron microscopy (Fig 3F and 3G). Collectively, these data suggest Delta-challenged mice have increased cellular responses and inflammation compared to Alpha-challenged K18-hACE-mice.
Fig 3. Histopathological and electron microscopy analysis of Alpha or Delta challenged K18-hACE2-mouse lungs.
20X images demonstrating inflammation in the lung tissue of Alpha and Delta SARS-CoV-2 challenged K18-hACE2 at Day 2 or 6 post-challenge (B) compared to no challenge (A). 20X images reveal margination at blood vessels within areas of inflammation (green arrows indicate counted marginating cells, red arrows indicate tissue-resident non-marginating cells)(C). Marginating inflammatory cells were counted within the tissue and were found to be increased in Delta challenge at Day 6 (Kruskal Wallis One-Way ANOVA, P = 0.0097) (D). Regional tissue inflammation was quantified as % total area of analyzed tissue (mm3) and was highest in Delta at Day 6 compared to no challenge (Kruskal Wallis One-Way ANOVA, P = 0.0012) (E). Error bars represent SD. Electron microscopy images displayed distribution of Delta virus particles in 1x103 PFU challenged mouse lung tissue (F,G). White arrows identify virions. Cell counts were reported for n = 5 mice per experimental group.
Pro-inflammatory cytokines are increased in mice challenged with Delta at day 6
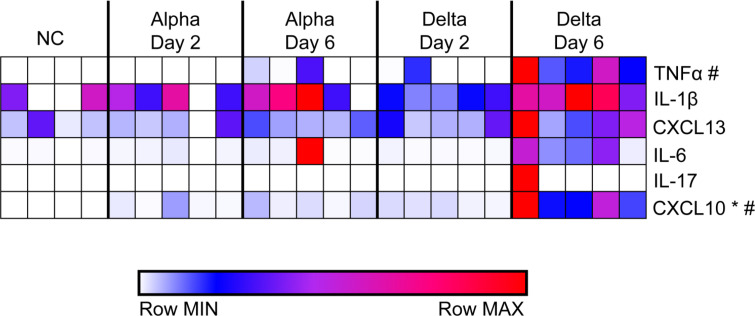
Histopathological analysis revealed significant inflammation in the lungs of Delta-challenged mice at day 6 compared to Alpha-challenged mice despite equal viral RNA burden. To better understand differences in the inflammatory profile of these tissues, pro-inflammatory cytokine levels in lung supernatant at day 2 or 6 were quantified to profile strain specific variations in the “cytokine storm” [47]. Insignificant amounts of cytokine were produced in the lungs of mice challenged with Alpha or Delta at day 2 (Fig 4). Alpha-challenged mice produced low amounts of IL-1β and CXCL13 compared to no challenge mice at day 2 or 6. By contrast, the cytokine response to Delta challenge at day 6 compared to no challenge revealed high levels of TNFα, and CXCL10 (One-Way ANOVA P = 0.0225, P = 0.0058) (Fig 4). CXCL10 was the only cytokine which statistical analysis proved to be higher in the lung supernatant of Delta-challenged mice than Alpha (One-Way ANOVA P = 0.0105). The inflammatory response within the lungs of mice challenged with Delta was elevated compared to Alpha. To gain insight into the specific mechanisms of the immunological host response, we performed RNAseq to characterize the total transcriptional profile of Alpha- and Delta-challenged K18-hACE2-mouse lungs.
Fig 4. Analysis of cytokines production in lung supernatants from Alpha or Delta challenged K18-hACE2 transgenic mice at 2- or 6-days post-challenge.
Non-challenged, Alpha- or Delta-challenged lung supernatants were used to determine local cytokine production in response to challenge. Concentrations (pg/mL) were graphed using Morpheus to reveal relative levels of cytokines compared to non-challenged lungs. * = statistical significance between Alpha day 6 and Delta day 6 (One-Way ANOVA P = 0.0105). # = statistical significance between no challenge and Delta Day 6 (One-Way ANOVA P = 0.0225 TNFα; P = 0.0058 CXCL10). (n = 5 except in NC where n = 4).
RNAseq analysis of lung tissues from mice challenged with Alpha or Delta shows no difference in viral sgRNA
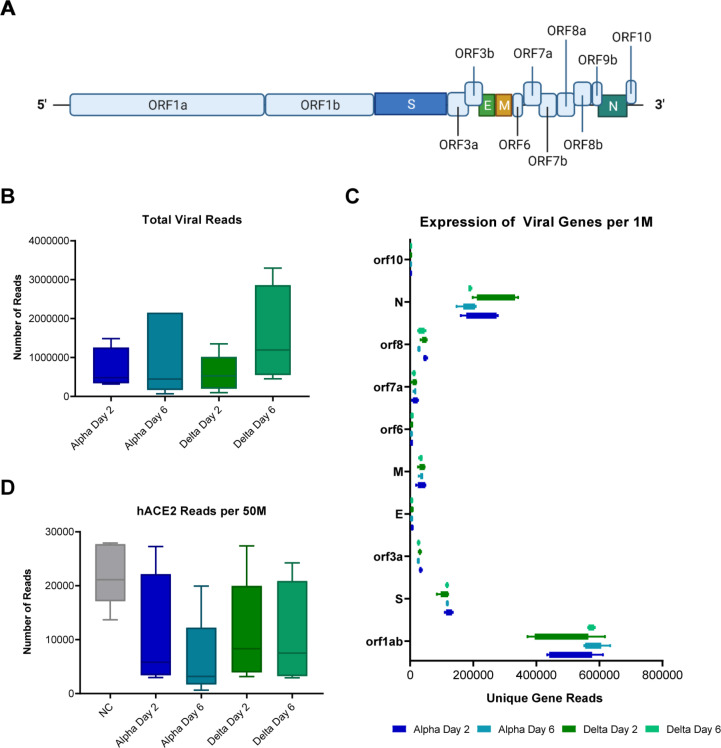
We utilized RNA that was isolated from the lung tissue of challenged and no challenge mice at 2- and 6-days post-challenge for Illumina transcriptomic analysis. RNA sequencing reads were first mapped to the SARS-CoV-2 viral genome to quantify the expression levels of viral genes over the course of infection (Fig 5A). This revealed no significant difference between the two strains (Fig 5B and 5C) which directly supported the qRT-PCR analysis (Fig 2). It is known that SARS-CoV-2 infection results in down-regulation of hACE2 expression [48, 49]. Therefore, we mapped RNA reads to the hACE gene to analysis this effect. Although not significant, a trend of lowered hACE2 expression occurred in Alpha challenged mice, whereas Delta challenged mice appeared to experience only slightly reduced hACE2 expression (Fig 5D). These observations together with qRT-PCR nucleocapsid data described above, suggest that with equal viral burden (same challenge dose and equal viral RNA burden), the differences in LD100 of Alpha and Delta as well as the disease phenotypes are likely related to differences in host response to VOC and not overall viral load.
Fig 5. RNAseq analysis of SARS-CoV-2 genes in challenged mouse lungs.
Schematic of the SARS-CoV-2 genome (A). Total viral read counts in each group (B) as well as quantification of viral reads per 1M RNAseq reads obtained (C). Reads of human ACE2 gene of the K18-hACE2-mice were quantified per 50M reads obtained per lung sample (D). (n = 4 in Alpha Day 2 and Delta Day 6; n = 5 in Alpha Day 6 and Delta Day 2).
Delta challenge causes a higher magnitude of host gene expression change compared to Alpha
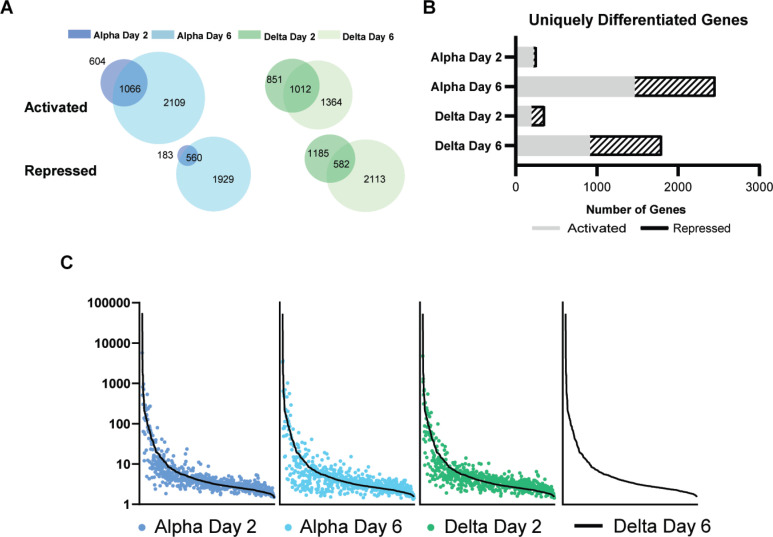
To compare the host responses of Delta- and Alpha-challenged mice, we mapped lung RNA sequencing reads to the mouse genome. We observed that Alpha or Delta challenge in K18-hACE2-mice result in distinct transcriptional changes. Two days after challenge with Alpha, 2,413 statistically significant genes (FDR P ≤ 0.04) were differentially expressed compared to no challenge mice, a difference which increases more than two-fold to 5,664 genes at day 6. In Delta groups, differential expression encompasses 3,048 and 4,489 genes respectively. Venn diagrams of the gene sets comparing by variant and time-points illustrate overlaps in gene sets that are either activated or repressed. Both activated and repressed gene sets increase from day 2 to 6 (Fig 6A). Further filtration of these sets of genes to identify those that are uniquely affected by VOC challenge and timepoint revealed that Alpha challenge results in more genes differentially regulated at day 6 than Delta challenge (Fig 6B). A set of core-activated genes (S2 Table) was identified through a four-way comparison of VOC and timepoint. Despite their different disease pathology, Alpha and Delta challenge activate a similar set of host genes with differences lying in the extent to which each gene is respectively regulated. Common genes which were activated by Alpha challenge at day 2 were also activated at day 6 and by Delta challenge at both times with different fold changes. Commonly repressed genes followed a similar trend. Together, these trends support that the nuances of the host response to Alpha and Delta infection may largely contribute to the differences that are seen in disease pathology.
Fig 6. Total transcriptomic analysis of mouse genes in non-challenged mice compared to Alpha and Delta challenged mice at days 2 and 6.
RNAseq reads from challenged mice were compared to non-challenged mice to determine differential gene expression (A). Genes that were statistically significant are indicated in relation to challenge day and their relative expression as being activated or repressed. Venn diagrams are shown to illustrate the amount of overlap in significant genes between days 2 and 6. The total number of unique differentially expressed genes are represented per each challenge day and relative expression (B). (n = 4 in Alpha Day 2 and Delta Day 6; n = 5 in Alpha Day 6 and Delta Day 2).
GO analysis revealed unique systems of enriched genes in mice challenged with Delta
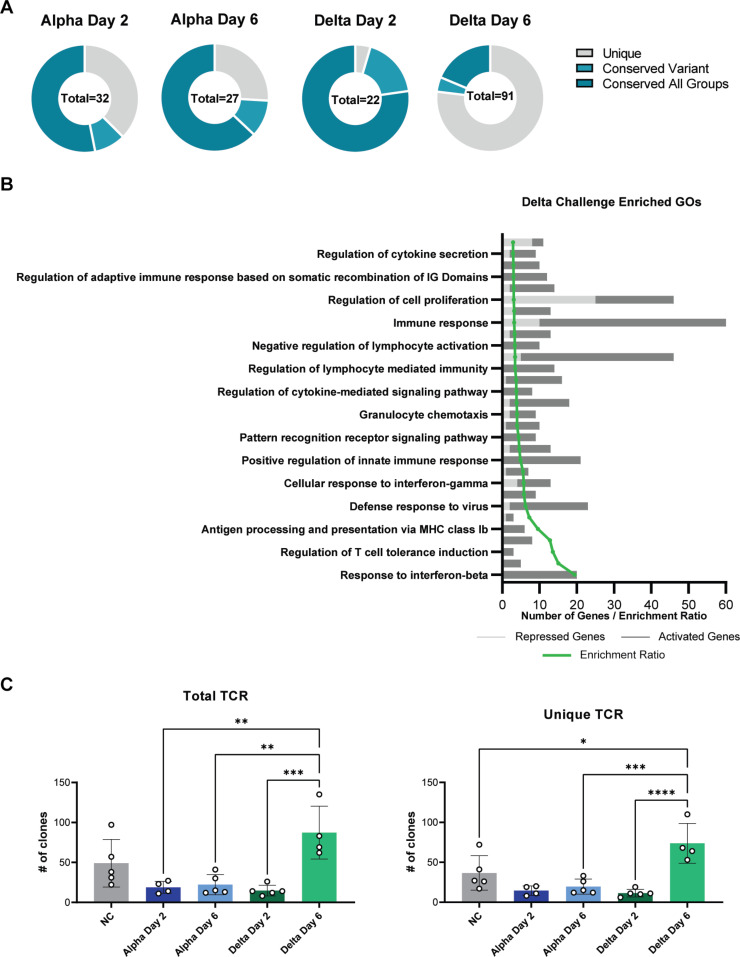
Gene Ontology (GO) term analysis was performed to systematically group transcriptomic observations to identify specific pathways and gene systems whose expression changes in response to Alpha and Delta challenge. Relative to Alpha-challenged mice, Delta-challenged mice had a higher number of unique GO terms that were enriched at day 6 (Fig 7A). To categorize the specifically enriched terms in Delta at day 6, pathway enrichment ratios were calculated for the top 30 GO terms (Fig 7B). The expression of genes that were annotated with GO terms pertaining to immune responses, anti-viral, or lymphocyte recruitment were increased, as were genes of pathways associated with T cells and responses to IFN-β (Fig 7B). To deepen the finding by GO term analysis that T cell response genes are enriched in Delta challenged mice, we performed T cell receptor (TCR) clonotype analysis using RNAseq reads. Within the no challenge control mice, we detected ~50 total clonotypes. In Alpha-challenged mice, the number of clones were decreased relative to no challenge at both day 2 and 6. By contrast, Delta-challenged mice had decreased clones at day 2, but we observed a 6-fold increase in TCR clones at day 6 (Fig 7C). In counts of unique TCR clonotypes, the same trends were observed: at day 6, Delta challenge had the most total unique TCR clones compared to non-challenged mice, day 6 post Alpha-challenged mice, and day 2 post Delta-challenged mice (Fig 7D). These data suggest that Delta may induce more robust T cell response.
Fig 7. Gene ontology analysis of systems of genes in challenged mice.
Circle plots indicate the number of unique or conserved sets of genes that were found to be enriched per each group (A). The top 30 enriched GO terms are shown and plotted per their relative level of expression and enrichment ratio (B). TCR clonotypes were mapped and represented as Total clones (C) or unique clones per unique CDR3 sequence (D). Total TCR clones (One-Way ANOVA P = 0.0005) and unique clones were increased between Delta day 2 and day 6 (One-Way ANOVA P<0.0001). (n = 4 in Alpha Day 2 and Delta Day 6; n = 5 in Alpha Day 6 and Delta Day 2).
Delta challenge causes increases interferon dependent gene expression
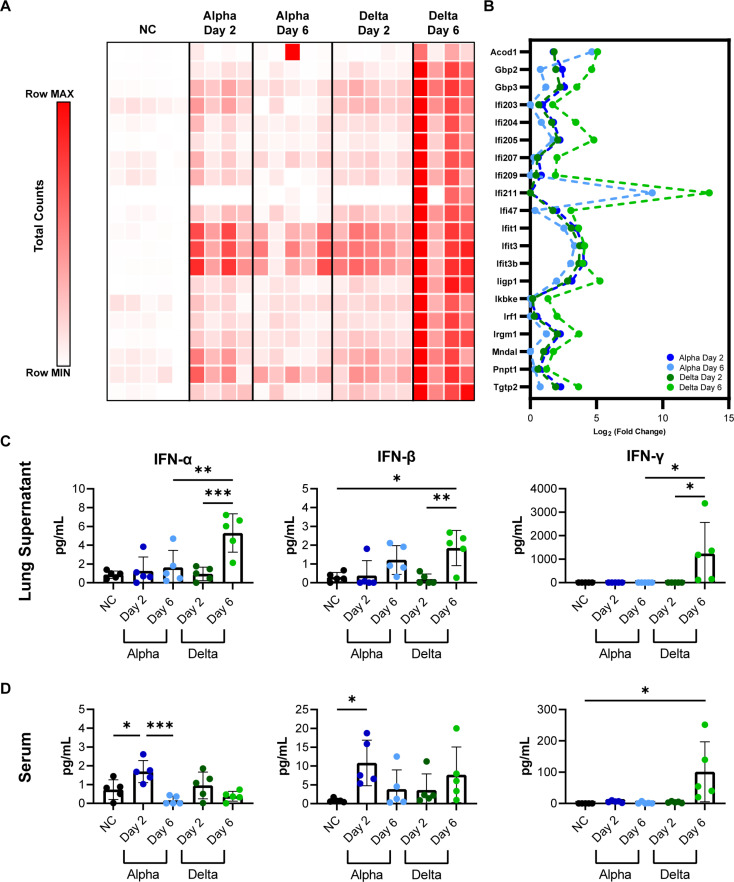
Gene expression analysis focused by GO terms revealed Delta challenge induced expression of interferon response genes (Fig 7). Interferon is an important regulator of the antiviral response in humans. It’s variable expression in response to specific VOC strains of SARS-CoV-2 exemplifies the intensity of differences in the host response. TGTP1 and IFI211, two interferon related genes, are the highest-expressed genes in Delta at day 6 at a fold change of 52,725 or 11,733 higher than in no challenge mice. In Alpha at day 6 these genes are 3,367 and 591-fold higher than no challenge. Due to the important role of interferons in the anti-viral response to SARS-CoV-2 which may be differentially engaged by VOC, we continued to compare the top 20 genes involved in general interferon responses. Total gene counts in lung RNA from each experimental group are shown in Fig 8A. Delta at day 6 shows high interferon dependent gene expression as well as the highest fold change difference (Fig 8B). Collectively, these data suggest Delta challenge results in higher interferon response. To test that hypothesis, we directly measured type I and II interferons in lung supernatants and serum. Modest production of IFN-α or IFN-β were observed in both lung and serum by both variants (IFN-α: P = 0.0008 Delta Day 6 vs No Challenge in lung supernatant; P = 0.0456 Alpha Day 2 vs No Challenge in serum; IFN-β: P = 0.0126 Delta Day 6 vs No Challenge in Lung Supernatant; P = 0.0474 Alpha Day 2 vs No Challenge in serum); however, Delta challenge resulted in higher IFN-γ at day 6 compared to day 2 and both Alpha challenge timepoints (P = 0.0265 in lung supernatant, P = 0.0103 in serum compared to no challenge) (Fig 8C and 8D). This dynamic production of interferon subtypes that appears to be unique to challenge strain and timepoint, speaks to the interesting differences in host response that may set apart variants in the K18-hACE2 SARS-CoV-2 challenge model. As variants continue to appear and be incorporated into current research projects, these phenotypic and pathological changes will need to be appreciated.
Fig 8. Analysis of interferon dependent gene expression and production of type I and type II interferon in lungs and serum of K18-hACE2-mice.
Total RNA counts per interferon dependent gene are shown per each mouse, time-point, and variant challenge group (A). Each square represents the total RNA counts per gene from one mouse. Interferon dependent genes are shown with relative fold change plotted (B). Interferon alpha, beta, and gamma production in lung supernatant (C) and serum (D) of K18-hACE2-mice. (n = 5).
Discussion
In this study, we modeled the pathogenesis of SARS-CoV-2 variants of concern in the K18-hACE2-mouse challenge model. Based on our observations that the Delta variant differs in disease manifestation from Alpha and Beta VOC-challenge, we performed a transcriptomic experiment to compare Alpha- and Delta-challenged mice at disease-significant timepoints. Despite a similar viral RNA burden in the respiratory tract at day 2 and 6, Delta causes more significant inflammation in the lungs than Alpha as infection progresses. The host response also varies over time, with Delta causing increased antiviral cytokine production, specifically IFN-γ, at day 6. GO term analysis of Delta-challenged mice suggested a greater number of immunological pathways that are implicated at time points in the variant’s pathogenesis compared to Alpha, further indicating the unique nature of the host response that is mounted against each individual variant of SARS-CoV-2.
Severe COVID-19 is perpetuated by the dysregulated production of pro-inflammatory cytokines, including type I and II interferons, in a “cytokine storm” pathology [47, 50, 51]. We observed that CXCL10, a chemokine that is produced downstream of IFN-γ signaling, in addition to IFN-α, IFN-β and IFN-γ were significantly higher in the lung supernatant of mice 6 days after Delta variant challenge, than in mice after Alpha challenge. RNAseq analysis corroborated these findings, with higher expression of interferon pathways genes in the lung tissue. The interferon signaling pathways, and the increased expression of interferon-stimulated genes (ISG) in COVID-19 patient lung tissues implicate the critical role of interferon in SARS-CoV-2 pathogenesis, but it has not been determined if the cytokines contribute more to suppressing viral replication, or augmenting the immune response through their dual pro- or anti-inflammatory abilities, thus contributing to greater disease [50, 51]. Delta challenge causes a significant increase in lung and serum IFNγ which is greater than what has been previously reported in K18-hACE2-mice using different challenge strains such as ancestral WA-1 [41, 44] IFNγ is a potent antiviral cytokine that is produced later in the timeline of viral challenge and often may not be observable in widely used mouse models of lethal SARS-CoV-2 challenge because of their short survival timelines [52]. IFNγ production is typically associated with the activation of natural killer cells, innate lymphoid cells, or Th1-like adaptive immune cells [45]. Its high concentrations in the lung of Delta mice could corroborate greater recruitment of these cell populations identified in histopathology data as infiltrating cells (Fig 3). Together the observations of this study suggest that Delta challenge causes enhanced innate immune responses compared to Alpha (Fig 9). Liu et al. in their comparison of Delta and an ancestral strain, examined cell infiltration by flow cytometry and immunofluorescence staining and confoundingly reported fewer inflammatory cells in Delta lungs than ancestral-strain despite very similar cytokine profiles between variants [53]. While our results alone only begin to suggest a differential engagement of interferons by Delta during its host pathogenesis, and combined with Liu et al.’s data show an abrogated cellular response between Delta and prior VOC strains, further comprehensive work would be necessary to fully map out the timelines of signaling mechanisms and cellular response profiles responsible for unique VOC pathology.
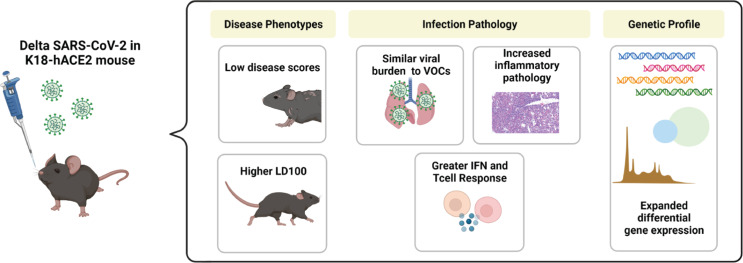
Fig 9. SARS-CoV-2 Delta variant challenge in the K18-hACE2 transgenic mouse model.
Male, 8 week-old K18-hACE2 transgenic mice were intranasally challenged with 103 PFU Delta SARS-CoV-2 and the host response to virus was analyzed at 2 and 6 days post-challenge. Delta challenge results in attenuated disease scores compared to previous VOC, and extended survival time at this dose. Despite similar viral RNA burdens, Delta-challenged mice experience greater inflammatory pathology in the lung with an expanded differential gene expression profile suggestive or greater interferon and T cell responses.
Surprisingly, we observed that the viral RNA burden of the respiratory tract was identical per day and per variant in the K18-hACE2-mice, as determined by both RT-qPCR and RNAseq. A clear question then remains: why does Delta induce a greater local inflammatory response in the lung? One study from Stolp, et al. demonstrated that SARS-CoV-2 variants differentially reduce the numbers of interferon-producing immune cells in K18-hACE2-mouse tissues, although it lacked the inclusion of Delta in its comparison of Alpha, Beta, Gamma, and ancestral strains [54]. Another possible mechanism that could be explored is the interaction of viral sgRNAs in regulating host gene expression. Open Reading Frames (ORFs) of SARS-CoV-2 have been implicated in controlling the host anti-viral response [55]. Studies report that the increase in viral burden due to Alpha leads to increases in some but not all sub-genomic RNAs from the genomic regions harboring variant-characteristic mutations [56]. Another study links the expression of sub-genomic RNAs in Alpha infection, specifically ORF9b, to antagonism of the host immune response and suppression of ISGs which would allow for more efficient viral replication and enhanced transmission [57]. Although viral gene reads in our analysis show no significant differences between Alpha and Delta sgRNAs, it is possible that our selected timepoints were too late to detect the differences previously reported at 10 and 24 hours post challenge, or that our challenge dose was too low. Still, our abundant transcriptomic data suggest an extensive list of gene pathways that were enriched during Delta but not Alpha infection.
We observed that 103 PFU challenge with Delta variant does not cause the dramatic weight loss and hypothermia reported in models of Alpha and Beta challenge [17]. One well known caveat of the K18-hACE2-mouse model is the fact that WA-1, Alpha and Beta strains enter the brain [44, 46, 58–64]. However, it appears that the Delta variant remains more localized in the lungs of challenged mice (Fig 3). At higher challenge doses (104 PFU), our lab and others have shown that viral RNA can be detectable in the brain as early as day 6 following Delta challenge [16, 43]. In data not yet published, we have also observed that at later time points, 103 PFU challenge with Delta can still result in some brain tissue burden, based on qRT-PCR analysis. It’s generally accepted that K18-hACE2-mice reach mortality due to brain infection, supported by manifestations of severe disease including hypothermia, and heightened inflammatory profiles. Seehusen et al. showed that Delta’s infectious potential in neurons is equal to that of previous VOC [65]. It is possible that a delay in the entry of Delta to the murine brain allows for an increased survival time and leaves more time for the development of severe infection and pneumonia in the lung. However, viral kinetics studies would be required to further measure this.
We appreciate that the K18-hACE2-mouse model has caveats to consider. The transgenic model expressing human ACE2 in addition to mouse ACE2 addresses SARS-CoV-2’s higher affinity for binding human ACE2, which reduces species tropism and allows virus to cause lethal disease in the mice. As is common in transgenic protein expression, hACE2 under the epithelial cell cytokeratin-18 (K18) promoter is not expressed with the identical tissue localization as it would be in humans. Therefore, viral localization should always be carefully considered. Our observations show that Delta accomplishes lung infection in the mouse and induces strong pneumonia. One popular alternative to the K18-hACE2-mouse model is the Syrian Golden Hamster. After SARS-CoV-2 challenge, hamsters develop more observable respiratory disease and pneumonia due to a higher affinity for hamster ACE2 [66, 67]. Delta has been reported to be highly pathogenic in golden hamsters, with increased tropism for the respiratory tract and low development of disease phenotypes like weight loss and drops in temperature [33, 68]. The similarities between mouse and hamster models provide evidence for Delta’s unique disease manifestation, defending the importance of its use in preclinical models when compared to prior strains.
With the emergence of the Omicron variant, it’s clear that novel variants of SARS-CoV-2 will continue to arise with enhanced disease capabilities. To continue preclinical COVID-19 research with small animal models, strains that more accurately recapitulate present human disease are necessary. Understanding the unique disease mechanisms of each VOC and the caveats they may present in these models will help to advance research into the identification or improvement of therapeutics.
The data presented here show that a sublethal challenge with Delta induces inflammation that is more histopathological than the Alpha VOC and increases interferon responses. This study also defines the doses of SARS-CoV-2 VOC that are useful for considering in performing preclinical studies. Our findings of enhanced pathogenesis from the Delta variant compared to the Alpha variant recapitulate some of the findings of Liu, et al. where the Delta variant at a higher challenge dose was found to engage the murine host immune response differently from an ancestral SARS-CoV-2 strain [16]. The use of RNAseq allowed us to characterize the entire transcriptome of the mouse lung to further understand the effect of challenge by either Alpha or Delta variants. We observed massive differential gene expression caused by Delta despite that fact that the mice did not reach mortality. Our study identified unexpected and uncharacteristic aspects of Delta’s disease pathogenesis. Together, our study outcomes underscore the need to continue to understand SARS-CoV-2 especially outside of the immunogenicity differences of the spike protein that are most often focused upon. We plan to build upon the findings of this study and characterize single-cell gene expression of the host response in a continued effort seeking to understand ways to combat this virus.
Materials and methods
Biosafety, animal, and ethics statement
K18-hACE2-mice purchased from Jackson Laboratory (B6.Cg-Tg(K18-ACE2)2Prlmn/J; JAX strain number #034860) were used for in vivo challenge studies under the approval of West Virginia University IACUC protocol number 2009036460. SARS-CoV-2 viral propagation work in addition to the challenge studies were conducted in West Virginia University’s High Containment Biosafety Laboratory Level 3 facility under the IBC protocol number 20-04-01. Tissue samples including SARS-CoV-2-challenged mouse serum and lung supernatant collected in BSL3 experiments were treated with 1% Triton (per volume, TRI Reagent (Zymo R2050-1), or formalin-fixedbefore exiting high containment for use in BSL2.
SARS-CoV-2 cultivation and K18-hACE2 mouse challenge
WA-1, Alpha, and Beta strains of SARS-CoV-2 were obtained from BEI: USA-WA-1/2020 (WA-1; BEI NR-52281) (GenBank accession number: MN985325), hCoV19/England/204820464/2020 (Alpha; NR-54000)(GISAID: EPI_ISL_683466), and hCoV19/South Africa/KRISP-EC-K005321/2020 (Beta; BEI NR-54008) (GISAID: EPI_ISL_678570). The SARS-CoV-2 Delta variant (B.1.617.2 hCoV-19/USA/WV-WVU-WV118685/2021) was obtained from a patient viral transport medium swab (GISAID Accession ID: EPI_ISL_1742834). All SARS-CoV-2 strains were propagated in Vero E6 cells (ATCC-CRL-1586) to prepare viral stocks. All viral strains were NGS sequenced to confirm the strains’ genomes. No mutations were identified that would suggest altered pathogenesis. For variant comparison studies, K18-hACE2 mice were anesthetized with an IP injection of ketamine (Patterson Veterinary 07-803-6637) /xylazine (Patterson Veterinary 07-808-1947) then intranasally challenged with a volume of 50 uL (25uL per nare) at: 103 PFU/dose (8 week old male and female received WA-1 or Beta; 12 week old female mice received Alpha; 13 week old female received Delta), or 104 PFU/dose (17 week old female received WA-1; 8 week old male and female received Alpha or Beta; 20 week old female received Delta). Mouse cohort details are summarized in S1 Table. For timepoint comparison study, male age-matched mice at 8 weeks old were infected with 103 PFU. Viral doses were prepared from the first passage collections from infected Vero E6 cells.
Disease score of SARS-CoV-2 challenged mice
K18-hACE2 mice were evaluated daily after viral challenge using in-person health assessments in the BSL3 as well as the SwifTAG Systems video monitoring until the planned euthanasia timepoint. Health assessments of the mice were scored based on the following criteria: weight loss (, appearance, activity, eye closure, and respiration using a scaling system that has been described previously [42, 43]. Each day, mice were awarded an additive score comprised of the five sub-scores. Mice that scored 5 or above on the health assessment or reached 20% weight loss required immediate euthanasia. For reporting purposes, the scores of each mouse in a group were combined each day and reported as the average.
Euthanasia and tissue collection
Mice were euthanized using an IP injection of Euthasol (390mg/kg) (Pentobarbital) followed by cardiac puncture. The blood from cardiac puncture was collected in BD Microtainer gold serum separator tubes (Catalog No: 365967), and spun down at 15,000 x g for 5 minutes to separate the serum. Nasal wash was performed by pushing 1mL of PBS through the nasal pharynx and collected in a microcentrifuge tube. 500μL of nasal wash was added to 500μL of TRI reagent for RNA purification and the remainder of the nasal wash was frozen and saved for future serological work. Lungs were dissected and separated: the right lobe of the lung was homogenized in 1mL of PBS in gentleMACS C tubes (order number: 130-096-334) using the m_lung_02 program on the gentleMACS Dissociator. For RNA purification, 300μL of lung homogenate was added to 1000μL of TRI Reagent (Zymo research) an additional 300 μL of lung homogenate was centrifuged at 15,000 x g for 5 minutes to collect the lung supernatant for cytokine analysis. The left lung was fixed using 10% neutral buffered formalin for histopathology. Whole brain tissue was also homogenized in 1mL PBS in gentleMACS C tubes using the same setting as lung on the gentleMACS Dissociator. For RNA purification from brain, 1000μL of TRI Reagent was added to 500μL of brain homogenate.
qPCR SARS-CoV-2 viral copy number analysis of lung, brain, and nasal wash
The Direct-zol RNA miniprep kit (Zymo Research R2053) was used to purify RNA from lung, brain, and nasal wash following the manufacturer’s protocol. SARS-CoV-2 copy numbers were assessed through qPCR using the Applied Biosystems TaqMan RNA to CT One Step Kit (Ref: 4392938) and a previously described reaction utilizing the nucleocapsid primers (F: ATGCTGCAATCGTGCTACAA; R: GACTGCCGCCTCTGCTC); and TaqMan probe (IDT:/56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/) synthesized according to Winkler. et al., 2020 [43, 44]. Purified RNA samples with a concentration less than 100ng/uL were not diluted for use in qPCR reactions. All Nasal Wash RNA samples were used at a set volume of 2 uL due to low RNA quantification Each sample reaction was prepared in triplicate and analyzed in MicroAmp Fast optical 96 well reaction plates (Applied Biosystems 4306737) on the StepOnePlus Real-Time System machine using the following parameters: Reverse transcription for 15 minutes at 48°C, activation of AmpliTaq Gold DNA polymerase for 10 minutes at 95°C, and 50 cycles of denaturing for 15 seconds at 95°C and annealing at 60°C for 1 minute.
Luminex and Meso Scale Discovery cytokine analysis
The R&D 5-plex mouse magnetic Luminex assay (Ref LXSAMSM) was used to quantify cytokines: IL-1β, CXCL13, TNFα, IL-6, IFN-γ, IL-17, and CXCL10 in lung supernatant. Manufacturer protocols were followed to prepare samples. Mouse cytokine plate was analyzed on the Luminex Magpix and concentration values (pg/mL) were calculated based off standard curves generated for each cytokine in the assay. IFN-α, IFN-β and IFN-γ were additionally quantified using MSD U-PLEX Interferon Combo 1 (ms) assay kit (Catalog No K15320K-1) and manufacturer protocols. MSD assay plates were analyzed using the Meso Scale Discovery Sector 2400.
Histology and electron microscopy
The left lobes of lungs were immediately fixed in 10mL of 10% neutral buffered formalin. Fixed lungs were paraffin embedded into 5μm sections. Sections were stained with hematoxylin and eosin and visualized on the DynamyxTM digital pathology platform. Lungs were scored for chronic and acute inflammation in the lung parenchyma, blood vessels, and airways by a blinded pathologist. Pulmonary inflammation was quantified by measuring the total area of lung tissue involved by inflammation. The predominant inflammatory cell type was noted. To quantify vascular margination of inflammatory cells, five representative arteries were identified within areas involved by inflammation. Total length of endothelium of these vessels was measured and the number of marginating inflammatory cells in the cross section were manually counted. Areas involved by inflammation were further evaluated by electron microscopic examination. The areas of interest were punched from the paraffin embedded tissue block and processed for electron microscopy. Ultrathin sections were cut on a Leica Ultra-Microtome, collected on copper mesh grids, stained using uranyl acetate and lead citrate and viewed using a Jeol 1010 electron microscope (FEI) with attached AMT camera.
Illumina library preparation, sequencing, and in silico bioinformatic analysis
After purification, the concentration of RNA in each sample was quantified using Qubit 3.0 Fluormeter using the RNA high sensitivity (Life Technologies) kit. RNA integrity was assessed using an Agilent TapeStation by Admera Health (South Plainfield, NJ). Illumina sequencing libraries were prepared after samples were DNAsed with KAPA RNA HyperPrep Kit with RiboErase (Basel, Switzerland). The resulting libraries passed standard Illumina quality control PCR tests and were sequenced using the Illumina NovaSeq s4 4000 at Admera Health. A total of ~100 million 150 base pair reads were acquired per sample. Sequencing data will be deposited to the Sequence Read Archive. The reads were trimmed for quality and mapped to the Mus musculus reference genome using CLC Genomics Version 21.0.5. Two mice were excluded due to no detectable viral reads (n = 5 NC, 4 Alpha Day 2, 5 Alpha Day 6, 5 Delta Day 2, 4 Delta Day 6). An exported gene expression browser table is available upon request. Statistical analysis of read data was performed with CLC’s Differential Expression for RNA Seq tool and genes were annotated with reference mouse gene ontology terms. Quantification of the number of activated or repressed genes unique to each experimental group was performed using Venny 2.1 [69]. Genes from each experimental comparison with significant fold changes (Bonferroni ≤ 0.04) were submitted to the WEB-based Gene SeT AnaLysis Toolkit’s Over Representation Analysis (ORA) software compared to the reference set “affy mg u74a” to determine GO terms from gene ontology and biological process databases (FDR ≤ 0.05) [70]. GO Term heat maps were generated using Morpheus [71]. Raw read data is available at NCBI SRA: PRJNA808079. An expression browser containing fold change and total counts data has been updated to Mendeley Data [72].
Statistical analyses
The statistical comparisons made in these studies were calculated using GraphPad Prism version 9. Statistical analyses were performed with n ≥ 3 for all K18-ACE2 mice studies. Comparisons of normally-distributed data sets were made using ordinary one-way ANOVA with Dunnett’s multiple comparisons test. Kruskal-Wallis with Dunn’s multiple comparisons tests were used to assess non-parametric distributed datasets. Kaplan-Meier survival curves were created, and Log-rank (Mantel-Cox) tests were used to test the significance of survival differences between sample groups.
Supporting information
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This project was supported by the Vaccine Development Center at the West Virginia University Health Sciences Center. We want to thank the WVU Flow Cytometry & Single Cell Core Facility for the use of their equipment. Figure preparation was supported by GraphPad Prism and BioRender.
We would like to acknowledge Mary Tomago-Chesney for sectioning and performing H&E staining on tissues. We would like to thank the Electron Microscopy, Histopathology and Tissue Bank Core for obtaining Electron Microscopy images of the delta variant in lungs. Lastly, we thank Drs. Laura Gibson and Clay Marsh for supporting this COVID-19 vaccine research.
Data Availability
All raw RNA sequencing fastq illumina read files are available at the NCBI SRA (bioproject accession number: PRJNA808079). Additional supporting information including CLC Genomics formatted expression browser containing all raw and statistically analyzed RNAseq data is available at: https://data.mendeley.com/datasets/jgtchwdh7x/1 (DOI:10.17632/jgtchwdh7x.1).
Funding Statement
Research Challenge Grant no. HEPC.dsr.18.6 from the Division of Science and Research, WV Higher Education Policy Commission. The MSD QuickPlex SQ120 in the WVU Flow Cytometry & Single Cell Core Facility is supported by the Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P30GM121322 (TME CoBRE) and P20GM103434 (INBRE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Galloway SE, Prabasaj P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. 2021;70(3):95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021. Apr 15;592(7854):438–43. doi: 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 3.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021. May 13;593(7858):266–9. doi: 10.1038/s41586-021-03470-x [DOI] [PubMed] [Google Scholar]
- 4.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science (80-). 2021. May 21;372(6544). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charmet T, Schaeffer L, Grant R, Galmiche S, Chény O, Von Platen C, et al. Impact of original, B.1.1.7, and B.1.351/P.1 SARS-CoV-2 lineages on vaccine effectiveness of two doses of COVID-19 mRNA vaccines: Results from a nationwide case-control study in France. Lancet Reg Heal—Eur. 2021. Sep;8:100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall VJ, Foulkes S, Saei A, Andrews N, Oguti B, Charlett A, et al. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet. 2021. May 8;397(10286):1725–35. doi: 10.1016/S0140-6736(21)00790-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinde V, Bhikha S, Hoosain Z, Archary M, Bhorat Q, Fairlie L, et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021. May 20;384(20):1899–909. doi: 10.1056/NEJMoa2103055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021. May 20;384(20):1885–98. doi: 10.1056/NEJMoa2102214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N Engl J Med. 2021;384(23):2187–201. doi: 10.1056/NEJMoa2101544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchings MDT, Ranzani OT, Dorion M, D’Agostini TL, de Paula RC, de Paula OFP, et al. Effectiveness of ChAdOx1 vaccine in older adults during SARS-CoV-2 Gamma variant circulation in São Paulo. Nat Commun [Internet]. 2021. [cited 2022 Jan 12];12(1). Available from: 10.1038/s41467-021-26459-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies NG, Jarvis CI, van Zandvoort K, Clifford S, Sun FY, Funk S, et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021. May 13;593(7858):270–4. doi: 10.1038/s41586-021-03426-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. Sars-cov-2 variant of concern 202012/01 has about twofold replicative advantage and acquires concerning mutations. Viruses [Internet]. 2021. Mar 1 [cited 2022 Jan 12];13(3). Available from: https://pubmed.ncbi.nlm.nih.gov/33804556/ doi: 10.3390/v13030392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patone M, Thomas K, Hatch R, Tan PS, Coupland C, Liao W, et al. Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: an observational cohort study. Lancet Infect Dis. 2021. Nov 1;21(11):1518–28. doi: 10.1016/S1473-3099(21)00318-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veneti L, Seppälä E, Storm ML, Salamanca BV, Buanes EA, Aasand N, et al. Increased risk of hospitalisation and intensive care admission associated with reported cases of SARS-CoV-2 variants B.1.1.7 and B.1.351 in Norway, December 2020–May 2021. Vol. 16, PLoS ONE. Public Library of Science; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grint DJ, Wing K, Houlihan C, Gibbs HP, Evans SJW, Williamson E, et al. Severity of Severe Acute Respiratory System Coronavirus 2 (SARS-CoV-2) Alpha Variant (B.1.1.7) in England. Clin Infect Dis [Internet]. 2021. Sep 6 [cited 2022 Jan 12]; Available from: https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab754/6365124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L, Liu Y, Tang X, He D. The Disease Severity and Clinical Outcomes of the SARS-CoV-2 Variants of Concern. Front Public Heal. 2021. Nov 30;9:1929. doi: 10.3389/fpubh.2021.775224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radvak P, Kwon HJ, Kosikova M, Ortega-Rodriguez U, Xiang R, Phue JN, et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun. 2021. Dec 1;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayarri-Olmos R, Johnsen LB, Idorn M, Reinert LS, Rosbjerg A, Vang S, et al. The alpha/b.1.1.7 sars-cov-2 variant exhibits significantly higher affinity for ace-2 and requires lower inoculation doses to cause disease in k18-hace2 mice. Elife [Internet]. 2021. Nov 1 [cited 2022 Jan 5];10. Available from: https://pubmed.ncbi.nlm.nih.gov/34821555/ doi: 10.7554/eLife.70002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Vol. 21, Nature Reviews Immunology. Nature Research; 2021. p. 626–36. doi: 10.1038/s41577-021-00592-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021. Aug 12;385(7):585–94. doi: 10.1056/NEJMoa2108891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS-CoV-2 variants of concern are emerging in India. Vol. 27, Nature Medicine. Nature Research; 2021. p. 1131–3. doi: 10.1038/s41591-021-01397-4 [DOI] [PubMed] [Google Scholar]
- 22.Nasreen S, Chung H, He S, Brown KA, Gubbay JB, Buchan SA, et al. Effectiveness of COVID-19 vaccines against variants of concern in Ontario, Canada. medRxiv [Internet]. 2021;2021.06.28.21259420. Available from: 10.1101/2021.06.28.21259420 [DOI] [Google Scholar]
- 23.Christensen PA, Olsen RJ, Wesley Long S, Subedi S, Davis J, Hodjat P, et al. Delta variants of SARS-CoV-2 cause significantly increased. medRxiv [Internet]. 2021. Oct 7; Available from: 10.1101/2021.07.19.21260808 [DOI] [Google Scholar]
- 24.Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N Engl J Med. 2021. Apr 15;384(15):1412–23. doi: 10.1056/NEJMoa2101765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell [Internet]. 2021. Jun 24 [cited 2022 Jun 10];184(13):3426–3437.e8. Available from: https://pubmed.ncbi.nlm.nih.gov/33991487/ doi: 10.1016/j.cell.2021.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe [Internet]. 2021. Mar 10 [cited 2022 Jan 12];29(3):463–476.e6. Available from: http://www.cell.com/article/S1931312821000822/fulltext doi: 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 2021 275. 2021. Mar;27(5):917–24. doi: 10.1038/s41591-021-01318-5 [DOI] [PubMed] [Google Scholar]
- 28.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. 2021. [cited 2021 Nov 30];596(7871):276–80. Available from: https://www.nature.com/articles/s41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature [Internet]. 2021. Mar 9; Available from: doi: 10.1038/s41586-021-04245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021. Apr 1;592(7852):116–21. doi: 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora P, Kempf A, Nehlmeier I, Graichen L, Sidarovich A, Winkler MS, et al. Delta variant (B.1.617.2) sublineages do not show increased neutralization resistance [Internet]. Vol. 18, Cellular and Molecular Immunology. Nature Publishing Group; 2021. [cited 2022 Jan 12]. p. 2557–9. Available from: https://www.nature.com/articles/s41423-021-00772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun [Internet]. 2021. Nov 1 [cited 2022 Jan 12];124:102715. Available from: /pmc/articles/PMC8354793/ doi: 10.1016/j.jaut.2021.102715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature [Internet]. 2021; Available from: 10.1038/s41586-021-04266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makdasi E, Zvi A, Alcalay R, Noy-Porat T, Peretz E, Mechaly A, et al. The neutralization potency of anti-SARS-CoV-2 therapeutic human monoclonal antibodies is retained against viral variants. Cell Rep. 2021. Sep 7;36(10). doi: 10.1016/j.celrep.2021.109679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunt AC, Case JB, Park YJ, Cao L, Wu K, Walls AC, et al. Multivalent designed proteins protect against SARS-CoV-2 variants of concern. bioRxiv Prepr Serv Biol [Internet]. 2021. Jul 7; Available from: http://www.ncbi.nlm.nih.gov/pubmed/34268509 doi: 10.1101/2021.07.07.451375 [DOI] [Google Scholar]
- 36.Stone S, Rothan HA, Natekar JP, Kumari P, Sharma S, Pathak H, et al. SARS-CoV-2 Variants of Concern Infect the Respiratory Tract and Induce Inflammatory Response in Wild-Type Laboratory Mice. Viruses [Internet]. 2021. Dec 24 [cited 2022 Jan 5];14(1):27. Available from: https://www.mdpi.com/1999-4915/14/1/27/htm doi: 10.3390/v14010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuai H, Chan JFW, Yuen TTT, Yoon C, Hu JC, Wen L, et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021. Nov 1;73. doi: 10.1016/j.ebiom.2021.103643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montagutelli X, Prot M, Levillayer L, Salazar EB, Jouvion G, Conquet L, et al. The B.1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 2021;1–16. [Google Scholar]
- 39.Moreau GB, Burgess SL, Sturek JM, Donlan AN, Petri WA, Mann BJ. Evaluation of K18-hACE2 Mice as a Model of SARS-CoV-2 Infection. Am J Trop Med Hyg. 2020;103(3):1215–9. doi: 10.4269/ajtmh.20-0762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz-Fontela C, Dowling WE, Funnell SGPP, Gsell PSS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19 [Internet]. Nature 2020. 586:7830 Nature Publishing Group; Sep 23, 2020 p. 509–15. Available from: https://www.nature.com/articles/s41586-020-2787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yinda CK, Port JR, Bushmaker T, Owusu IO, Purushotham JN, Avanzato VA, et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021;17(1):1–21. doi: 10.1371/journal.ppat.1009195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong TY, Horspool AM, Russ B, Ye C, Lee KS, Winters M, et al. Evaluating Antibody-Mediated Protection against Alpha, Beta, and Delta SARS-CoV-2 Variants of Concern in K18-Human ACE2 Transgenic Mice [Accepted Jan 2, 2022 PMCID: PMC Journal—In Process]. J Virol. 2022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong TY, Lee KS, Russ BP, Horspool AM, Kang J, Winters MT, et al. Intranasal administration of BReC-CoV-2 COVID-19 vaccine protects K18-hACE2 mice against lethal SARS-CoV-2 challenge. npj Vaccines [Internet]. 2022. Dec 14 [cited 2022 Mar 25];7(1):36. Available from: https://pubmed.ncbi.nlm.nih.gov/35288576/ doi: 10.1038/s41541-022-00451-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol [Internet]. 2020. Aug 24 [cited 2021 Aug 8];21(11):1327–35. Available from: https://www.nature.com/articles/s41590-020-0778-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun. 2020;11(1). doi: 10.1038/s41467-020-19891-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong W, Mead H, Tian L, Park JG, Garcia JI, Jaramillo S, et al. The K18-hACE2 Transgenic Mouse Model Recapitulates Non-Severe and Severe COVID-1 19 in Response to Infectious Dose of SARS-CoV-2 Virus 2 3. 2021. [cited 2022 Jan 11]; Available from: https://www.worldometers.info/coronavirus/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Vol. 11, Frontiers in Immunology. Frontiers Media S.A.; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klouda T, Hao Y, Kim H, Kim J, Olejnik J, Hume AJ, et al. Interferon-alpha or -beta facilitates SARS-CoV-2 pulmonary vascular infection by inducing ACE2. Angiogenesis. 2021; doi: 10.1007/s10456-021-09823-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020; doi: 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKechnie JL, Blish CA. The Innate Immune System: Fighting on the Front Lines or Fanning the Flames of COVID-19? Vol. 27, Cell Host and Microbe. Cell Press; 2020. p. 863–9. doi: 10.1016/j.chom.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type i interferon signaling. J Exp Med. 2020. Aug 4;217(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol. 2018. Sep 11;9(SEP):2061. doi: 10.3389/fimmu.2018.02061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Mostafavi H, Ng WH, Freitas JR, King NJC, Zaid A, et al. The Delta SARS-CoV-2 Variant of Concern Induces Distinct Pathogenic Patterns of Respiratory Disease in K18-hACE2 Transgenic Mice Compared to the Ancestral Strain from Wuhan. MBio [Internet]. 2022. Apr 14 [cited 2022 Jun 9]; Available from: https://journals.asm.org/journal/mbio [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stolp B, Stern M, Ambiel I, Hofmann K, Morath K, Gallucci L, et al. SARS-CoV-2 variants of concern display enhanced intrinsic pathogenic properties and expanded organ tropism in mouse models. Cell Rep. 2022. Feb 15;38(7). doi: 10.1016/j.celrep.2022.110387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci U S A [Internet]. 2021. Jun 8 [cited 2022 Jan 5];118(23). Available from: https://pubmed.ncbi.nlm.nih.gov/34021074/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker MD, Lindsey BB, Shah DR, Hsu S, Keeley AJ, Partridge DG, et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv [Internet]. 2021. Mar 4 [cited 2022 Jan 9];2021.03.02.433156. Available from: https://www.biorxiv.org/content/10.1101/2021.03.02.433156v2 [Google Scholar]
- 57.Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, Pelin A, et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv Prepr Serv Biol [Internet]. 2021. Jun 7; Available from: http://www.ncbi.nlm.nih.gov/pubmed/34127972 doi: 10.1101/2021.06.06.446826 [DOI] [Google Scholar]
- 58.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, et al. Lethal Infection of K18- hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus. J Virol [Internet]. 2007. Jan 15 [cited 2021 Aug 8];81(2):813–21. Available from: https://pubmed.ncbi.nlm.nih.gov/17079315/ doi: 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carossino M, Montanaro P, O’Connell A, Kenney D, Gertje H, Grosz KA, et al. Fatal neuroinvasion of SARS-CoV-2 in K18-hACE2 mice is partially dependent on hACE2 expression. bioRxiv Prepr Serv Biol [Internet]. 2021. Jan 15 [cited 2022 Jan 5]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/33469581 [Google Scholar]
- 60.Fumagalli V, Ravà M, Marotta D, Lucia P Di, Laura C, Sala E, et al. Controlled administration of aerosolized SARS-CoV-2 to K18-hACE2 transgenic mice uncouples respiratory infection and anosmia from fatal neuroinvasion. bioRxiv [Internet]. 2021. Aug 6 [cited 2022 Jan 5];2021.08.06.455382. Available from: https://www.biorxiv.org/content/10.1101/2021.08.06.455382v1 [Google Scholar]
- 61.Kumari P, Rothan HA, Natekar JP, Stone S, Pathak H, Strate PG, et al. Neuroinvasion and encephalitis following intranasal inoculation of sars-cov-2 in k18-hace2 mice. Viruses [Internet]. 2021. Jan 1 [cited 2022 Jan 5];13(1). Available from: https://pubmed.ncbi.nlm.nih.gov/33477869/ doi: 10.3390/v13010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chia PY, Ong SWX, Chiew CJ, Ang LW, Chavatte JM, Mak TM, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clin Microbiol Infect [Internet]. 2021; Available from: 10.1016/j.cmi.2021.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kissler SM, Fauver JR, Mack C, Olesen SW, Tai C, Shiue KY, et al. Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLoS Biol. 2021. Jul 1;19(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marc A, Kerioui M, Blanquart F, Bertrand J, Mitjà O, Corbacho-Monné M, et al. Quantifying the relationship between sars-cov-2 viral load and infectiousness. Elife. 2021. Sep 1;10. doi: 10.7554/eLife.69302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seehusen F, Clark JJ, Sharma P, Bentley EG, Kirby A, Subramaniam K, et al. Neuroinvasion and Neurotropism by SARS-CoV-2 Variants in the K18-hACE2 Mouse. Viruses [Internet]. 2022. [cited 2022 Jun 9];14(5):1020. Available from: https://www.mdpi.com/1999-4915/14/5/1020/htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. 2020. [cited 2022 Jan 26];583(7818):834–8. Available from: https://www.nature.com/articles/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. 2020. Jul 14 [cited 2022 Jan 5];117(28):16587–95. Available from: www.pnas.org/cgi/doi/10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohandas S, Yadav PD, Shete A, Nyayanit D, Sapkal G, Lole K, et al. Sars-cov-2 delta variant pathogenesis and host response in syrian hamsters. Viruses. 2021. Sep 1;13(9). doi: 10.3390/v13091773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olivers JC. Venny. An interactive tool for comparing lists with Venn’s diagrams [Internet]. Available from: http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 70.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res [Internet]. 2019;47(W1):W199–205. Available from: http://www.webgestalt.org. doi: 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morpheus [Internet]. [cited 2021 Oct 12]. Available from: https://software.broadinstitute.org/morpheus/
- 72.Lee K, Damron F. Transcriptomic Analysis of Alpha and Delta SARS-CoV-2 VOC in K18-hACE2 mice [Internet]. Vol. 1. Mendeley Data; 2022. [cited 2022 Aug 12]. Available from: https://data.mendeley.com/datasets/jgtchwdh7x/1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All raw RNA sequencing fastq illumina read files are available at the NCBI SRA (bioproject accession number: PRJNA808079). Additional supporting information including CLC Genomics formatted expression browser containing all raw and statistically analyzed RNAseq data is available at: https://data.mendeley.com/datasets/jgtchwdh7x/1 (DOI:10.17632/jgtchwdh7x.1).