Figure 2.
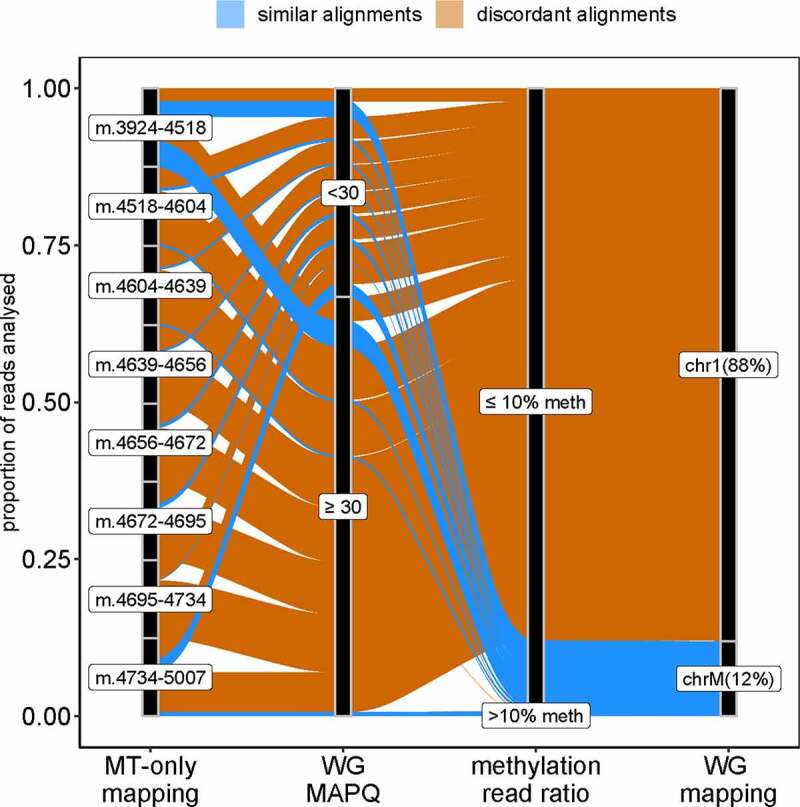
Differences in read mapping between MT-only and WG-alignment strategies.
The alluvial plot traces the mapping of the reads mapped with MT-only on the positions covered by less than 1,000 reads in WG-alignment. Concordant read alignment is shown in blue and discordant alignment is shown in orange. The majority (88%) of the 8,082 reads mapped on chromosome 1 (chr1) and only 12% mapped on mtDNA (chrM) in the WG-alignment. MAPQ: mapping quality determined using Bismark (Phred score). Methylation read ratio: ratio of non-converted cytosines to converted cytosines in any given read. WG-alignment: chromosome where the reads mapped with the WG-alignment strategy.
