Abstract
Multisystem inflammatory syndrome in children (MIS-C) is a severe complication of SARS-CoV-2 infections that occurs in the pediatric population. We sought to characterize T cell responses in MIS-C compared to COVID-19 and pediatric hyperinflammatory syndromes.
MIS-C was distinct from COVID-19 and hyperinflammatory syndromes due to an expansion of T cells expressing TRBV11–2 that was not associated with HLA genotype. Children diagnosed with MIS-C, but who were negative for SARS-CoV-2 by PCR and serology, did not display Vβ skewing. There was no difference in the proportion of T cells that became activated after stimulation with SARS-CoV-2 peptides in children with MIS-C compared to convalescent COVID-19. The frequency of SARS-CoV-2-specific TCRs and the antigens recognized by these TCRs were comparable in MIS-C and COVID-19.
Expansion of Vβ11–2+ T cells was a specific biomarker of MIS-C patients with laboratory confirmed SARS-CoV-2 infections. Children with MIS-C had robust antigen-specific T cell responses to SARS-CoV-2.
Keywords: Multisystem inflammatory syndrome in children (MIS-C), Coronavirus disease 2019 (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), T cell receptor (TCR) repertoire, Pediatrics
1. Introduction
Multisystem inflammatory syndrome in children (MIS-C) emerged as a complication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children during the first wave of the COVID-19 pandemic [1,2]. MIS-C is notable for several remarkable features, which distinguish this syndrome from COVID-19 in pediatric patients. While children with acute SARS-CoV-2 infections are often asymptomatic or present with mild upper respiratory symptoms, patients with MIS-C have prominent fevers, mucocutaneous involvement, and gastrointestinal symptoms [[3], [4], [5]]. The characteristic rashes, conjunctival injection, and mucositis of MIS-C is reminiscent of Kawasaki disease (KD) and toxic shock syndrome (TSS) [1,2]. MIS-C is further defined by its severity and a significant proportion of patients present in shock and require admission to the intensive care unit (ICU) [[3], [4], [5], [6]]. Curiously, MIS-C manifests about a month after the primary SARS-CoV-2 infection in children who mount an appropriate antibody response to the virus [3,4,[7], [8], [9], [10]].
The delayed presentation of MIS-C in comparison to the primary viral infection suggests that the adaptive immune response plays a role in driving the pathology of this disorder. While the majority of children with MIS-C are seropositive for antibodies that neutralize SARS-CoV-2, less is known about T cell responses [2,[5], [6], [7], [8], [9], [10]]. Circulating T cells are decreased in patients with MIS-C but display features of lymphocytes that are activated and proliferating, particularly in CD8+ T cells that can patrol the vasculature [[10], [11], [12]]. CD4+ and CD8+ T cells from patients with MIS-C display increased usage of the T cell Receptor Beta Variable 11–2 gene (TRBV11–2) that is not found in pediatric COVID-19, severe adult COVID-19, pre-pandemic KD, or TSS [[12], [13], [14], [15], [16]]. This expansion of Vβ11–2+ T cells resolves quickly with immunomodulatory treatment and is not found in convalescent MIS-C [14,16]. Further, greater V-beta (Vβ) repertoire skewing is associated with higher levels of circulating inflammatory cytokines and increased disease severity in patients with MIS-C [[13], [14], [15], [16]]. Porritt et al. and Sacco et al. demonstrated associations between HLA class I alleles (A02, C35, C04) and increased usage of TRBV11–2 in T cells from MIS-C patients; however, this has not been replicated in other studies [12,[14], [15], [16]]. Furthermore, it is not known if such TRBV11–2 expansions occur in other pediatric hyperinflammatory syndromes such as systemic juvenile idiopathic arthritis (sJIA) or macrophage activation syndrome (MAS).
To understand T cell responses in MIS-C, we evaluated the T cell receptor (TCR) repertoire and characteristics of SARS-CoV-2-specific T cells in these patients compared to children and adults with COVID-19 and other pediatric patients with hyperinflammatory syndromes. Our results confirm the previously observed enrichment of T cells expressing TRBV11–2 in MIS-C and further demonstrate that this expansion of Vβ11–2+ T cells is not found in other hyperinflammatory conditions of childhood such as KD, sJIA, and MAS. The increased usage of TRBV11–2 in this cohort was not associated with HLA type. SARS-CoV-2-specific T cell responses were evaluated through T cell assays and TCR sequencing. The proportion of T cells that responded to stimulation with SARS-CoV-2 peptides, the frequency of SARS-CoV-2-specific TCRs, and the distribution of viral antigens recognized by these SARS-CoV-2 specific TCRs were similar in patients with MIS-C and children with COVID-19.
2. Materials and methods
2.1. Patients and samples
Patients were recruited from Boston Children's Hospital (BCH) and provided a peripheral blood sample for the study. Children with MIS-C and COVID-19 were enrolled in the Taking on COVID-19 Together study or the Kawasaki Disease (KD) Biorepository between 5/2020–4/2021. MIS-C patients were included if they met the Centers for Disease Control (CDC) and/or World Health Organization (WHO) Case Definitions for MIS-C [[17], [18]]. Severe MIS-C was defined by admission to the intensive care unit or development of coronary artery aneurysms (CAAs) (z-score ≥ 2.5). Children with COVID-19 presented with fever, respiratory symptoms, and/or a COVID-19 exposure along with a positive SARS-CoV-2 test by polymerase chain reaction (PCR). Blood samples obtained 2 weeks after a positive PCR for SARS-CoV-2 were classified as convalescent COVID-19.
In addition, children with complete or incomplete KD (American Heart Association criteria), sJIA (modified International League of Associations for Rheumatology criteria, without the arthritis requirement), and MAS (2016 Classification Criteria) were also studied [[19], [20], [21]]. Children with KD unrelated to SARS-CoV-2 had no known close contacts with COVID-19 and tested negative for SARS-CoV-2 by both serology and PCR. Children with fever not related to SARS-CoV-2 who were evaluated in the Emergency Department, served as febrile controls. Pediatric controls with non-inflammatory musculoskeletal symptoms were recruited from the Rheumatology Clinic. Adult healthy controls also participated in the study.
The electronic medical record (EMR) was reviewed by a pediatric rheumatologist (LAH) to gather the necessary clinical data, which was entered into a REDCap database. Written informed consent and assent (when appropriate) were provided by the study subjects under the following Institutional Review Board (IRB) protocols: IRB—P00035409, X10-01-0308, and IRB—P00005723.
2.2. Cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood samples by Ficoll density gradient centrifugation and cryopreserved in liquid nitrogen.
2.3. T cell receptor sequencing
Genomic DNA was extracted from PBMCs (QIAamp Mini Blood DNA kit, Qiagen). A multiplex PCR was used to amplify the TCRβ complementarity determining region 3 (CDR3β), using a standard quantity of DNA. Sequencing was performed with the Illumina HiSeq platform (Adaptive Biotechnologies). Correction for PCR amplification bias was previously determined by using a complete, synthetic TCR repertoire to establish an amplification baseline and adjust the assay chemistry to minimize primer bias [22]. Residual PCR bias was addressed by using barcoded, spiked-in synthetic templates to assess sequence coverage. Resulting data were filtered and clustered using the relative frequency ratio between similar clones and a modified nearest neighbor algorithm to merge closely related sequences and remove both PCR and sequencing errors, as described [22,23].
The International ImMunoGeneTics (IMGT) system was used to align the sequences to a reference genome [24]. Subsequently, productive sequences were evaluated with the immunoSEQ™-Analyzer set of online tools. For diversity measures, the raw immunoSEQ dataset was imported in Python v3.7.0 using the import_adaptive_file() function in the tcrdist3 library. The data was then filtered to exclude clonotypes whose CDR3β sequence length was <4 amino acids and did not start with the consensus C amino acid or end with the consensus W/F amino acid. The productive frequency of each clonotype was then recalculated to account for excluded clones. The number of templates (copies) of each remaining clonotype was then used as input for plotting rarefaction curves and estimating Shannon Entropy at a fixed sample coverage of 0.2 using iNEXT, to identify potential outliers and correct for differences in sample size and sequencing depth in clonality estimates [25]. Using this approach, 3 patients were excluded from the diversity analysis due to inadequate sequencing depth (pediatric control, n = 1; MIS-C, n = 1; MAS, n = 1). Shannon entropy was used to assess repertoire diversity [26]. Clonality was measured by the median frequency of the 100 most abundant T cell clonotypes [27]. VDJ usage and V-J gene pairing were also determined. Chord diagrams were generated with R package circlize and heatmaps were created with GraphPad-Prism version 9 [28]. The primary TCR sequencing data are available at: clients.adaptivebiotech.com/pub/lam-2022-ci.
The immunoSEQ dataset and multiplexed identification of antigen-specific T cell receptors assay (MIRA) dataset from the ImmuneCODE database were sourced to provide TCR repertoires from adults with COVID-19 and SARS-CoV-2-specific TCRs, respectively [[29], [30], [31]]. Briefly, the immunoSEQ dataset is composed of TCRβ sequences from the adults (whole blood or PBMCs) with the following: 1) exposure to SARS-CoV-2 within the prior 2 weeks; 2) active COVID-19 based on a physician diagnosis or positive PCR test; 3) recovered COVID-19 defined by resolution of symptoms, 2 negative PCR tests, or clearance by a physician. Severe COVID-19 was defined as an adult with COVID-19 and a WHO ordinal severity scale rating of ≥6. In MIRA, SARS-CoV-2-specific peptide pools were used to identify antigen-specific T cells. The TCRs of these antigen-specific T cells were then sequenced. By using different combinations of antigen peptide pools, the target peptide recognized by a given TCR can be determined.
2.4. HLA typing
DNA was extracted from either whole blood or PBMCs (QIAamp Blood Midi/Mini Kit, Qiagen) from children with MIS-C SARS-CoV-2pos and pediatric COVID-19. High resolution sequencing of 11 HLA loci (A, B, C, DRB1/3/4/5, DQA1, DQB1, DPA1, DPB1) was performed by the HLA Research Testing Services of the American Red Cross. HLA allele frequencies in children with SARS-CoV-2 infections were compared to those in the National Marrow Donor Program, which contains HLA typing on millions of individuals in the United States [32].
2.5. SARS-CoV-2 peptide pool assays
PBMCs were stimulated with either water (negative control) or peptide pools covering the N-terminal S1 domain of the spike (S), nucleocapsid (N), and membrane (M) proteins of SARS-CoV-2 (Milteny Biotec) for 16 h. The cells were harvested and stained for CD154 (CD40 ligand, eBioscience) as a measure of TCR activation. CD154 expression was evaluated by flow cytometry.
2.6. Statistical analysis
One-way ANOVA with correction for multiple comparisons (Turkey correction) or unpaired, two-tailed t-tests were used to compare study groups. For TRBV gene usage analysis, two-way ANOVA with Dunnett's multiple comparison test was performed to compare study group and the frequency of each Vβ gene. Spearman's non-parametric rank correlation was used to test for a monotonic association between two variables. Statistical analysis was performed with GraphPad-Prism version 9 or R, including the package immunarch version 0.6.7 [33]. We ran the geneUsage function from immunarch to calculate gene segment frequencies for each TRBV gene per sample. We used Wilcoxon test to compare TRBV11–2 gene usage of patients with MIS-C SARS-CoV-2pos versus ImmuneCODE groups. Principal component analysis on TRBV gene usage fractions was performed using the prcomp() function from the stats package without centering and scaling the data. To assess whether gene usage frequency of TRBV11–2 was dependent on read depth sequencing, we down sampled clones per repertoire to the size of the smallest repertoire using the repSample function from immunarch and we calculated gene usage as described previously. To test whether the viral antigen profile recognized by SARS-CoV-2-specific TCRs was more similar between children with MIS-C and COVID-19 than expected by chance, we calculated the sum of differences in frequency of antigen-specific TCRs for each position of the viral genome between the two groups (observed), and then performed 1 million permutations of the genomic positions, re-calculating the sum of differences for each position at each iteration, creating an expected null distribution.
3. Results
3.1. Study cohorts
Children with MIS-C (n = 24), COVID-19 (n = 10), KD (n = 5), sJIA (n = 7), and MAS (n = 3) were studied along with pediatric febrile controls (n = 6), healthy children (n = 7), and adult controls (n = 5) (Tables 1 , S1-S3).
Table 1.
Clinical Characteristics of the Study Subjects.
| MIS-C |
Peds COVID-19 |
KD |
sJIA |
MAS |
Peds Febrile Controls |
Peds Controls |
Adult Controls |
|
|---|---|---|---|---|---|---|---|---|
| n = 24 |
n = 10 |
n = 5 |
n = 7 |
n = 3 |
n = 6 |
n = 7 |
n = 5 |
|
| Age-yrs (median, IQR) | 9.0, 6.3–13.8 | 11.0, 1.2–13.5 | 7.0, 1.5–10.5 | 6.0, 5.0–16.0 | 1.67, 1.2–12.0 | 4.5, 1.9–9.0 | 9.0, 6.0–10.0 | 26.5,25.0–28.8 |
| Sex (#, % female) | 15, 63% | 7, 70% | 2, 40% | 3, 43% | 2, 67% | 2, 34% | 4, 57% | 4, 80% |
| Race & Ethnicity (#, %) | ||||||||
| White, non-Hispanic | 4, 17% | 2, 20% | 3, 60% | 5, 71% | 2, 67% | 1, 17% | 2, 29% | 2, 40% |
| Black, non-Hispanic | 4, 17% | 0, 0% | 0, 0% | 0, 0% | 0, 0% | 0, 0% | 1, 14% | 0, 0% |
| Hispanic | 9, 38% | 5, 50% | 1, 20% | 1, 14% | 0, 0% | 3, 50% | 1, 14% | 0, 0% |
| Asian | 0, 0% | 1, 10% | 1, 20% | 0, 0% | 0, 0% | 1, 17% | 0, 0% | 1, 20% |
| Other | 2, 8% | 0, 0% | 0, 0% | 1, 14% | 1, 33% | 0, 0% | 0, 0% | 0, 0% |
| Unknown | 5, 21% | 2, 20% | 0, 0% | 0, 0% | 0, 0% | 1, 17% | 3, 43% | 2, 40% |
3.2. Children with SARS-CoV-2 infections
Patients with MIS-C met either the CDC or WHO case definitions for MIS-C (Table S1) [[17], [34]]. Fever was present in 100% of patients with MIS-C and gastrointestinal (GI) symptoms (92%), conjunctivitis (71%), and rash (38%) were also frequent. Children with MIS-C demonstrated evidence of systemic inflammation (median CRP 16.6 mg/dL, IQR 6.9–26.0), lymphopenia (median absolute lymphocyte count 0.92 × 10 [3]/mL, IQR 0.42–1.72) and coagulopathy (median D-dimer 3.1 μg/mL, IQR 1.4–6.2). Approximately 80% of MIS-C patients had positive SARS-CoV-2 serologies. Five patients tested negative for SARS-Co-V-2 by PCR and antibody levels but met the CDC or WHO Case Definitions for MIS-C and were thought to have MIS-C by their treating providers. Of the total MIS-C cohort, 11/24 (46%) were considered to have severe disease defined by admission to the intensive care unit (ICU) and/or development of CAAs. At the time of sampling, 79% of MIS-C patients had received immunomodulatory therapy, most commonly IVIG and/or glucocorticoids.
Most children with COVID-19 in this cohort had mild disease: 40% of patients were admitted to the hospital, 20% were treated with supplemental oxygen or non-invasive mechanical ventilation, and 10% required ICU level care (Table S1). Children with COVID-19 were frequently febrile (70%) with respiratory symptoms (60%), and all were positive for SARS-CoV-2 by PCR. In contrast to MIS-C, children with COVID-19 mostly lacked GI and mucocutaneous symptoms.
3.3. Pediatric hyperinflammatory syndromes
To compare MIS-C to other pediatric hyperinflammatory syndromes, pediatric patients with KD, sJIA, and MAS were also studied. Of the 5 children with KD, 60% met criteria for complete KD and 40% had CAAs (Table S2). None of the patients with KD presented with shock or were admitted to the ICU. All children with sJIA and MAS had active disease with fever, rash, and/or elevated inflammatory markers (Table S3).
3.4. TCR repertoire diversity and clonality in MIS-C
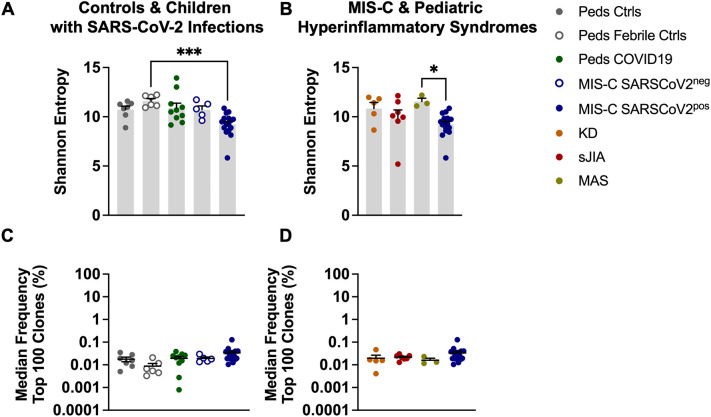
Shannon entropy was used to quantify TCR repertoire diversity in children with MIS-C compared to pediatric controls and pediatric COVID-19 (Fig. 1A) [26]. Children with MIS-C had the lowest levels of repertoire diversity but a statistically significant difference was noted only when comparing MIS-C patients with confirmed SARS-CoV-2 infection to pediatric febrile controls (Fig. 1A) (p ≤ 0.001). Children diagnosed with MIS-C who tested negative for SARS-CoV-2 by PCR and antibody tests (MIS-C SARS-CoV-2neg), were grouped separately from those children who had laboratory evidence of infection (MIS-C SARS-CoV-2pos) to determine if there were differences in TCR repertoire characteristics across these two groups. There were no significant differences in Shannon entropy in the MIS-C SARS-CoV-2pos vs. MIS-C SARS-CoV-2neg patients. Next, we compared the TCR repertoire diversity of children with MIS-C to other pediatric hyperinflammatory syndromes (Fig. 1B). There was a trend towards reduced diversity in the MIS-C patients that only reached statistical significance when compared to patients with MAS (Fig. 1B) (p ≤ 0.05). TCR repertoire clonality was measured by determining the median frequency of the 100 most abundant T cell clones in each sample. There were no differences in T cell clonal expansion across the study groups (Fig. 1C-D, S1). In summary, these findings suggest that children with MIS-C have a slight decrease in TCR repertoire diversity but no major T cell clonal expansions.
Fig. 1.
TCR Repertoire Diversity and Clonality in MIS-C. A-B) Shannon Entropy, a measure of repertoire diversity, estimated at fixed sample coverage. C—D) Median frequency (percent) among the top 100 most frequent clones with T cell clone defined as amino acid sequence in CDR3β of the TCR. Summary data on bar graphs is mean ± standard error. Statistical testing for A-D: Kruskal-Wallis test with Dunn's multiple comparisons test. P value * ≤ 0.05, *** ≤ 0.001. CDR3, complementarity determining region 3; TCR, T cell receptor; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrls, controls; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome.
3.5. Increased usage of TRBV11–2 in T cells from children with MIS-C
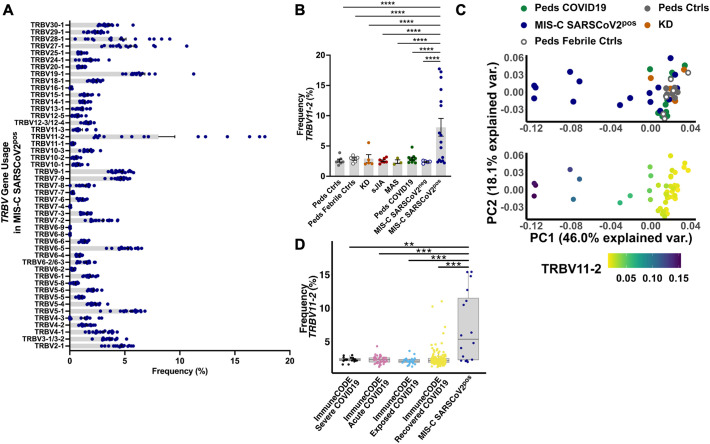
As described previously, T cells that express the TRBV11–2 gene in the CDR3β domain of the TCR were markedly increased in children with MIS-C compared to pediatric controls and pediatric COVID-19 (Fig. 2A, B) (p ≤ 0.0001) [[12], [13], [14], [15], [16]]. Skewed usage of TRBV11–2 was also significantly different in patients with MIS-C compared to children with other hyperinflammatory syndromes (KD, sJIA, MAS) (Fig. 2B) (p ≤ 0.0001). This finding was also confirmed by a principal components analysis (PCA) of TRBV gene usage. MIS-C SARSCoV-2pos patients were separated from the remaining study groups by PC1, which explained 46% of the variance and included TRBV11–2 as the main driver of PC1 (Fig. 2C) (Spearman rho = −0.88, P < 2.2e-16). Interestingly, children who met the case definition for MIS-C and were diagnosed with MIS-C by their treating providers but tested negative for SARS-CoV-2 did not display such Vβ skewing (Fig. 2B). Given this difference in MIS-C SARS-CoV-2pos vs. MIS-C SARS-CoV-2neg patients, we continued to analyze these groups separately.
Fig. 2.
Expansion of T cells expressing TRBV11–2 in MIS-C. A) The frequency of T cell clonotypes expressing each TRBV gene in patients with MIS-C SARSCoV2pos. B) The frequency of T cell clonotypes expressing TRBV11–2 in each study group. Statistical testing: two-way ANOVA comparing study group and Vβ gene with the Dunnett's multiple comparisons test. P value **** ≤ 0.0001. C) Principal components analysis (PCA) of TRBV gene usage. D) The frequency of T cell clonotypes expressing TRBV11–2 in MIS-C SARSCoV2pos and adults in the ImmuneCODE database. Statistical testing: Wilcoxon test. Summary data on bar graphs is mean ± standard error in A-B and median with IQR in D. P value, **** ≤ 0.0001, *** = 0.001, ** = 0.01, * = 0.05.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome; var., variance.
Next, we leveraged the ImmuneCODE database to compare TRBV11–2 usage in children with MIS-C SARS-CoV-2pos and adults with severe acute COVID-19 (n = 19), acute COVID-19 (n = 40), recovered COVID-19 (n = 159), or known SARS-CoV-2 exposure (n = 26). T cell clonotypes expressing TRBV11–2 were significantly more frequent in MIS-C SARS-CoV-2pos patients compared to all of the adult groups evaluated in ImmuneCODE, even after controlling for read depth differences between the datasets (Fig. 2D, S2A). The frequency of Vβ11–2+ T cells did not differ in adults based on the severity of COVID-19 illness. A small number of adults (4/159) in the COVID-19 recovered group had TRBV11–2 expansions, defined as a frequency >2 standard deviations (SD) above the mean.
There was a bimodal distribution of TRBV11–2 usage in MIS-C SARS-CoV-2pos patients. In some children, the frequency of T cell clonotypes expressing TRBV11–2 was comparable to controls (n = 6) while others had profound expansions of these T cells (n = 10) (Fig. 2B). TRBV11–2 expansions were defined as frequencies >2 SD above the mean of pediatric controls. We sought to determine what factors may explain this distribution of TRBV11–2 expression in MIS-C SARS-CoV-2pos patients. There was a non-statistically significant trend towards increased frequencies of Vβ11–2+ T cells in children with severe MIS-C, defined by admission to the ICU or development of CAAs (Fig. S2B). The number of children in the MIS-C SARS-CoV-2pos group who provided a blood sample prior to immunomodulatory treatment was small (n = 3), thus limiting the comparison of Vβ11–2+ T cell pre- vs. post-treatment (Fig. S2C). There was no statistically significant correlation between TRBV11–2 usage in T cells and patient age, body mass index (BMI), or days from admission to biosample collection (Fig. S2D—F).
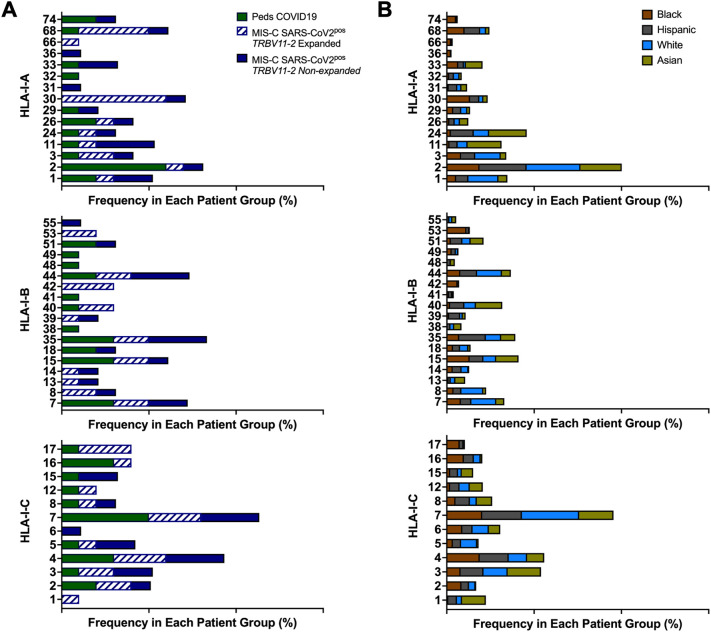
In some prior studies, expansions of Vβ11–2+ T cells in children with MIS-C were associated with HLA class I alleles, specifically A02, B35, and C04 [15,16]. Through HLA typing, the frequencies of HLA alleles in children with MIS-C SARS-CoV-2pos and COVID-19 were compared to the National Marrow Donor Program (NMDP), which contains HLA typing on millions of individuals in the United States [32]. Children with MIS-C SARS-CoV-2pos did not demonstrate increased usage of HLA-I A02, B35, or C04 compared to children with COVID-19 and the larger population represented in the NMDP registry (Fig. 3 ). There appeared to be an increased usage of HLA-I A30 in children with MIS-C and expanded Vβ11–2+ T cells; however, this allele is more commonly found in Black and Hispanic compared to White and Asian individuals in the United States based on the NMDP registry (Fig. 3B). Given the increased incidence of MIS-C in Black and Hispanic children, the observed enrichment of HLA-I A30 may reflect these racial and ethnic differences. No other HLA class I or II associations were noted in MIS-C patients with expanded T cells that expressed TRBV11–2 (Fig. 3, S3 and Table S4).
Fig. 3.
HLA Class I Alleles in MIS-C. A) The frequency of each HLA class I genotype in children with pediatric COVID-19 and MIS-C. B) The of frequency each HLA class I genotype in the National Marrow Donor Program database shown by race and ethnicity as defined by US census categories.
Peds, pediatric; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children.
Children with MIS-C SARS-CoV-2pos had decreased usage of TRBV7–2 and TRBV28–1 compared to some study groups, but these effect sizes were much less profound compared to the differences document in TRBV11–2 (Fig. S2G—H) and not consistent across all study groups.
Thus, increased expression of TRBV11–2 in T cells was specific to MIS-C patients and was not found in children or adults with COVID-19 or pediatric patients with hyperinflammatory syndromes. Children in the MIS-C SARS-CoV-2neg did not have expansions of Vβ11–2+ T cells, suggesting distinct T cell responses in patients who tested positive for SARS-CoV-2 by either PCR or serology.
3.6. Diverse TRBJ gene usage in MIS-C
In patients with MIS-C, there was no observed skewing of T Cell Receptor Beta Joining (TRBJ) genes in the TCR (Fig. 4A). The usage of TRBJ genes was similar across all study groups (Fig. 4A). In Vβ11–2+ T cells from MIS-C SARS-CoV-2pos patients, pairing with Jβ genes remained polyclonal and similar to children with COVID-19 (Fig. 4B, C).
Fig. 4.
Diverse TRBJ Gene Usage in MIS-C: A) Heatmap depicting the mean frequency of T cell clonotypes expressing each TRBJ gene in the given study groups. B—C) Chord diagram depicting the Vβ-Jβ pairing of total T cell clonotypes expressing TRBV11–2 in B) children with COVID-19 (n = 10) and C) MIS-C SARSCoV2pos patients (n = 16). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome.
3.7. SARS-CoV-2-specific T cells in MIS-C
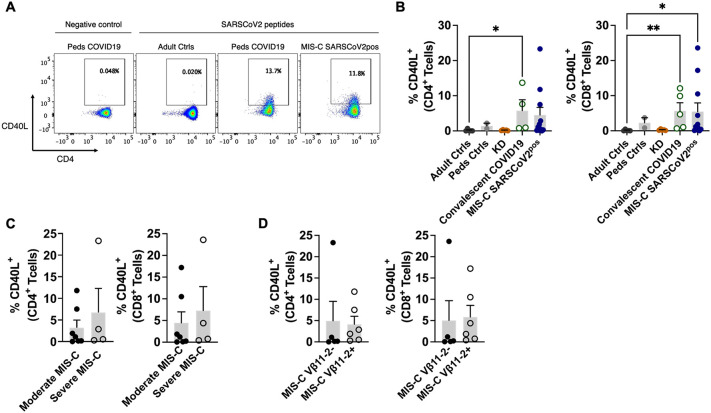
We next sought to characterize SARS-CoV-2-specific T cells in patients with MIS-C. Since MIS-C occurs several weeks after an initial SARS-CoV-2 infection, this group was compared to children with convalescent COVID-19. After stimulation with peptide pools covering the S1, N, and M viral proteins, antigen-specific T cells were identified by their expression of CD154 (CD40 ligand), a T cell activation marker. The frequency of CD4+ and CD8+ T cells that recognized SARS-CoV-2 in patients with MIS-C SARS-CoV-2pos did not differ significantly from children with convalescent COVID-19 (Fig. 5A, B). Similarly, there was no statistical difference in the frequency of CD40L+ T cells in mild vs. severe MIS-C SARS-CoV-2pos or in MIS-C patients with vs. without expansions of T cells expressing TRBV11–2 (Fig. 5C, D). These results suggested that MIS-C patients had similar frequencies of SARS-CoV-2-specific T cells as children with convalescent COVID-19.
Fig. 5.
SARS-CoV-2-Specific T Cells in MIS-C. Peripheral blood mononuclear cells were incubated with either water (negative control) or peptide pools covering the S1, N, and M viral proteins. SARS-CoV2-antigen-specific T cells were identified by their expression of CD154 (CD40 ligand). A) Flow cytometry dot plots depicting CD40L expression on T cells from a healthy control, child with convalescent COVID-19, and child with MIS-C SARS-COV-2pos. B) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in the given study group. C) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in children with moderate or severe MIS-C SARS-COV-2pos. D) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in MIS-C SARS-COV-2pos patients with and without expansions of T cell clonotypes expressing TRBV11–2. Statistical testing: Kruskal-Wallis test with Dunn's multiple comparisons test for panel B and for Mann Whitney tests for panels C—D. P value * ≤ 0.05, ** ≤ 0.01.
Summary data on bar graphs is mean ± standard error.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; KD, Kawasaki disease.
3.8. SARS-CoV-2-specific T cell receptors in MIS-C
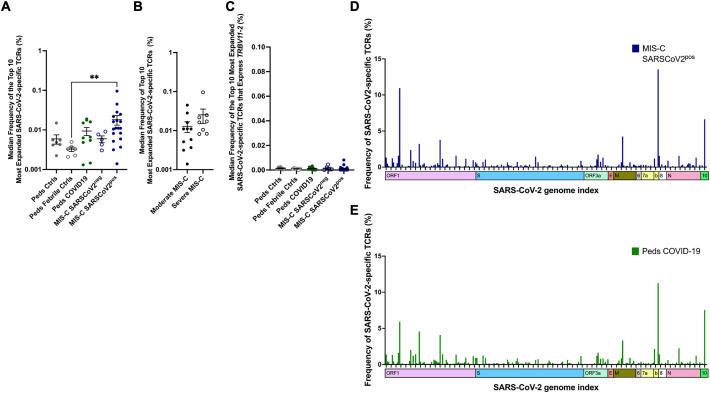
Using the MIRA dataset, we evaluated SARS-CoV-2-specific TCRβ sequences in children with MIS-C. There was a trend towards expanded SARS-CoV-2-specific TCRs in the MIS-C SARS-CoV-2pos patients; however, this difference only reached statistical significance when compared to febrile controls (p = 0.02) (Fig. 6A). There was a trend towards increased frequency of SARS-CoV-2-specific TCRs in patients with severe vs. moderate MIS-C that did not reach statistical significance (Fig. 6B). TCRs that expressed TRBV11–2 and recognized SARS-CoV-2 were not enriched in MIS-C patients (Fig. 6C). The distribution of viral antigens recognized by SARS-CoV-2-specific TCRs were similar in children with MIS-C and COVID-19 (Fig. 6D-E) (permutation test). There was an increase in frequency of TCRs that recognize ORF1 of SARS-CoV-2 in MIS-C SARS-CoV-2pos patients; however this difference was driven primarily by expanded TCRs in only 2 of the MIS-C patients. Thus, characteristics of SARS-CoV-2-specific TCRs were similar in children with MIS-C and COVID-19.
Fig. 6.
SARS-CoV-2-Specific T Cell Receptors in MIS-C. SARS-CoV-2-specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs for each patient in each study group. Statistical analysis Kruskal-Wallis test with Dunn's multiple comparison test. B) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs in patients with moderate and severe MIS-C. Statistical analysis: Mann-Whitney test C) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs that express TRBV11–2 in each patient and each study group. Statistical analysis Kruskal-Wallis test with Dunn's multiple comparison test. D-E) Viral antigens recognized by SARS-CoV-2-specific TCRs in D) MIS-C SARSCOV2pos and E) pediatric COVID-19. Statistical analysis by permutation test with no difference between MIS-C SARSCOV2pos and peds COVID-19.
P value, ** ≤ 0.01.
Summary data on bar graphs is mean ± standard error.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; TCR, T cell receptor.
4. Discussion
We sought to characterize T cell responses in MIS-C compared to COVID-19 and pediatric hyperinflammatory syndromes by leveraging high-throughput TCR sequencing and T cell immune assays. The most distinctive feature of the TCR repertoire in patients with laboratory-confirmed MIS-C was the skewed usage of TRBV11–2. In addition, children with MIS-C had robust antigen-specific T cell responses to SARS-CoV-2 that were comparable to pediatric patients with COVID-19.
The expansion of T cell clonotypes expressing TRBV11–2 documented in our cohort of children with MIS-C adds to the substantial literature supporting this observation [[12], [13], [14], [15], [16]]. Previously, increased usage of TRBV11–2 was found to be unique to patients with MIS-C compared to patients with COVID-19, KD, TSS, and pediatric febrile illnesses [[14], [15], [16]]. Our results expand upon these findings and demonstrate that MIS-C is also differentiated from sJIA and MAS by this Vβ skewing. By leveraging the publicly available ImmuneCODE database, we also show that increased TRBV11–2 usage is not observed in a large number of adults with active COVID-19, regardless of disease severity; however, it is occasionally seen in adults who have recovered from the infection. Interestingly, increased Vβ11–2+ T cells were not observed in the 5 children who met case definitions for MIS-C but tested negative for SARS-CoV-2 by PCR and serology. These patients presented during the first wave of the COVID-19 pandemic when the epidemiologic link to SARS-CoV-2 infection was relatively easy to establish. They were also diagnosed with MIS-C by their treating providers. The lack of skewed Vβ usage in the MIS-C SARS-CoV-2neg patients suggests that these children may have presented with an alternate condition that mimicked the findings typically observed in MIS-C. If true, quantifying the frequency of Vβ11+ T cells, which can be achieved relatively quickly by flow cytometry, may prove to be a useful diagnostic tool for MIS-C.
An expansion of T cells expressing TRBV11–2 was observed in more than half of the patients with MIS-C in this study. Others have also observed that Vβ skewing is not universal in MIS-C [[12], [13], [14]]. Factors associated with an increased frequency of Vβ11–2+ T cells include severe MIS-C and high levels of inflammation [[13], [14], [15], [16]]. In our cohort, there was a trend towards increased TRBV11–2 usage in patients with severe disease compared to moderate MIS-C, which did not reach statistical significance. Sacco et al. and Moreews et al. found that immunomodulatory treatment led to rapid contraction of Vβ11–2+ T cells in patients with MIS-C [14,16]. Thus, treatment-related effects may have contributed to the bimodal distribution of Vβ gene usage that was observed in this study, although the paucity of pre-treatment samples did not permit a robust assessment. Finally, some studies have demonstrated HLA class I associations with TRBV11–2 expansions in MIS-C, while other reports have not reproduced these findings [12,14,15]. To date, our cohort of 19 MIS-C patients with HLA typing represents the largest group of patients with such data and we did not observe HLA class I or II associations with TRBV11–2 expansions.
The functional significance of TRBV11–2 expansions in MIS-C remains to be determined. Vβ and Jβ genes are spliced together to create diversity in the antigen recognition domain of TCRβ. Our study and others show that the TRBV11–2 gene in MIS-C is paired with polyclonal Jβ genes [12,[14], [15], [16]]. Prior studies have leveraged single cell RNA sequencing (scRNA-seq) to confirm that there is no bias in the pairing of TCRβ chains expressing TRBV11–2 with specific TCRα chains [12]. These TCR repertoire features indicate a non-clonal expansion of a particular Vβ gene and are suggestive of superantigen-mediated pathology. Indeed, computational modeling indicates that the sequence and structure of the SARS-CoV-2 spike protein has features of a superantigen and TRBV11–2 may interact with this superantigen-like motif [15,35]. Further, flow cytometry and scRNA-seq show that Vβ11–2+ T cells are activated and pro-inflammatory [12,14,16]. Yet, these findings are descriptive with no functional studies confirming the role of Vβ11–2+ T cells in MIS-C. The delay in the presentation of MIS-C compared to the primary SARS-CoV-2 infection is not typical of diseases driven by superantigens, although some reports indicate that SARS-CoV-2 may persist in some hosts outside of the respiratory tract [16,36]. Superantigens are typically HLA class II restricted. HLA associations, including those observed in our cohort, have not been clearly demonstrated in MIS-C. Further, the TRBV11–2 expansions observed in adults with convalescent COVID-19 raises questions about the pathogenicity of these T cells. A MIS-C-like illness (MIS-A) can occur in adults, but it is even more rare than in children. It seems unlikely that 4/159 patients with TRBV11–2 expansions in the ImmuneCODE database developed such a syndrome. Thus, it seems reasonable to conclude that adults can have increased frequencies of Vβ11–2+ T cells without hyperinflammation. It is also possible that this dissociation occurs in the pediatric population, although we did not have sufficient number of children with convalescent COVID-19 to explore this possibility.
We utilized two approaches to explore antigen-specific T cell responses in MIS-C. First, we characterized the frequency of T cells activated by stimulation with SARS-CoV-2 peptide pools. These T cells occurred at similar frequencies in patients with MIS-C SARS-CoV-2pos and children with convalescent COVID-19. Further, the frequency of these antigen-specific T cells did not correlate with disease severity or TRBV11–2 expansions. Second, we leveraged the publicly available MIRA dataset to identify SARS-CoV-2-specific TCRs. There was a non-statistically significant trend towards expanded SARS-CoV-2 TCRs in children with MIS-C SARS-CoV-2pos, particularly in children with severe MIS-C. Further studies in a larger number of children are needed to determine if the frequency of SARS-CoV-2-specific TCRs is associated with disease severity. The distribution of viral antigens recognized by these TCRs was similar in MIS-C SARS-CoV-2pos and pediatric COVID-19. There was no evidence that TCRs that express TRBV11–2 compared to other Vβ genes were more likely to recognize SARS-CoV-2. In total, children with MIS-C generated SARS-CoV-2-specfic T cells at levels that were comparable to children with COVID-19. In keeping with our prior findings that TRBV11–2 expansions are non-clonal, T cells expressing this Vβ gene were not enriched in the SARS-CoV-2-specific T cell subset.
Our work furthers the understanding of T cell responses in MIS-C; however, we acknowledge limitations to our study. While the number of MIS-C patients (n = 24) in our study is sizable compared to many other published cohorts, these children were recruited from a single center, which may limit generalizability. Since children with MIS-C can decompensate quickly and develop shock, they were treated rapidly at our center, often in the emergency department, with intravenous immunoglobulin (IVIG) and glucocorticoids. Thus, many children with MIS-C in this cohort had received immunomodulatory treatment prior to sampling. Arguably, the TCR repertoire is impacted less by such treatment compared to studies that evaluate protein and gene expression in T cells. Finally, antigen-specific T cell responses were identified by T cell assays and SARS-CoV-2 specific TCRs found in a publicly available dataset; however, we cannot absolutely confirm that these T cells recognize SARS-CoV-2 in vivo.
5. Conclusions
Expansions of Vβ11–2+ T cells was a specific biomarker of MIS-C patients with laboratory-confirmed SARS-CoV-2 infections and was not found in children and adults with COVID-19 or pediatric patients with hyperinflammatory syndromes. Children with MIS-C appeared to generate appropriate antigen-specific T cell responses to SARS-CoV-2, and these antigen-specific T cells were not associated with TRBV11–2 expansions.
Funding
This work was supported by the National Institutes of Health (NIAMS K08 AR073339, NIAMS P30 AR070253), the Rheumatology Research Foundation (Career Development Bridge Funding Award), and the Arthritis National Research Foundation (All Arthritis Grant). Adaptive Biotechnologies provided no-charge sequencing through Adaptive Antigen Map. The authors had full access to the raw data and analyzed and interpreted the data independently.
Declaration of Competing Interest
LAH has received salary support from CARRA; consulting fees from Sobi, Pfizer, and Adaptive Biotechnologies; and investigator-initiated research grants from Bristol-Myers Squibb.
Acknowledgements
We thank the patients and volunteers for participation in this study.
MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; COVID-19, coronavirus disease 2019; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2022.109106.
Appendix A. Supplementary data
The supplementary data includes supplementary tables 1-3 with additional clinical characteristics of the study chorts. The HLA typing information is provided in supplementary table 4. Supplementary figures 1-3 are also included.
References
- 1.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdoni L., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldstein L.R., et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dufort E.M., et al. Multisystem inflammatory syndrome in children in New York State. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whittaker E., et al. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated With SARS-CoV-2. Jama. 2020 doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belhadjer Z., et al. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 7.Gruber C.N., et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020 doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rostad C.A., et al. Quantitative SARS-CoV-2 Serology in children with multisystem inflammatory syndrome (MIS-C) Pediatrics. 2020;146 doi: 10.1542/peds.2020-018242. [DOI] [PubMed] [Google Scholar]
- 9.Consiglio C.R., et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183:968–981. doi: 10.1016/j.cell.2020.09.016. e967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter M.J., et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 2020 doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- 11.Vella L.A., et al. Deep immune profiling of MIS-C demonstrates marked but transient immune activation compared to adult and pediatric COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abf7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoste L., et al. TIM3+ TRBV11-2 T cells and IFNgamma signature in patrolling monocytes and CD16+ NK cells delineate MIS-C. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramaswamy A., et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–1095. doi: 10.1016/j.immuni.2021.04.003. e1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreews M., et al. Polyclonal expansion of TCR Vbeta 21.3(+) CD4(+) and CD8(+) T cells is a hallmark of multisystem inflammatory syndrome in children. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abh1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porritt R.A., et al. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J. Clin. Invest. 2021;131 doi: 10.1172/JCI146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacco K., et al. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat. Med. 2022 doi: 10.1038/s41591-022-01724-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention, Emergency preparedness and response: health alert network, 2020. Accessed June 16, 2022.
- 18.World Health Organization, Multisystem inflammatory syndrome in children and adolescents with COVID-19, 2020. Accessed June 16, 2022. https://www.who.int/publications/i/item/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19.
- 19.McCrindle B.W., et al. Diagnosis, treatment, and long-term management of kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135:e927–e999. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 20.Ravelli A., et al. 2016 classification criteria for macrophage activation syndrome complicating systemic juvenile idiopathic arthritis: a European League Against Rheumatism/American College of Rheumatology/Paediatric Rheumatology International Trials Organisation Collaborative Initiative. Arthritis Rheum. 2016;68:566–576. doi: 10.1002/art.39332. [DOI] [PubMed] [Google Scholar]
- 21.Petty R.E., et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J. Rheumatol. 2004;31:390–392. [PubMed] [Google Scholar]
- 22.Carlson C.S., et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 23.Robins H.S., et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh T.C.M., K.H., Chao, A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol. Evol. 2013;7:1451–1456. [Google Scholar]
- 26.Stewart J.J., et al. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 1997;34:1067–1082. doi: 10.1016/s0161-5890(97)00130-2. [DOI] [PubMed] [Google Scholar]
- 27.Pielou E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. [Google Scholar]
- 28.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 29.Nolan S., et al. A large-scale database of T-cell receptor beta (TCRbeta) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. Res. Sq. 2020 doi: 10.21203/rs.3.rs-51964/v1. [DOI] [Google Scholar]
- 30.Klinger M., et al. Combining next-generation sequencing and immune assays: a novel method for identification of antigen-specific T cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klinger M., et al. Multiplex identification of antigen-specific T cell receptors using a combination of immune assays and immune receptor sequencing. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiers M., Gragert L., Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 33.ImmunoMind Team . 2019. Immunarch: An R Package for Painless Bioinformatics Analysis of T-Cell and B-Cell Immune Repertoires. [Google Scholar]
- 34.World Health Organization Multisystem inflammatory syndrome in children and adolescents with COVID-19. May 15, 2020. https://www.who.int/publications/i/item/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19 [Accessed June 16, 2020]
- 35.Cheng M.H., et al. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yonker L.M., et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Invest. 2021;131 doi: 10.1172/JCI149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary data includes supplementary tables 1-3 with additional clinical characteristics of the study chorts. The HLA typing information is provided in supplementary table 4. Supplementary figures 1-3 are also included.